













| General Information: |
|
| Name(s) found: |
gi|34365725
[NCBI NR]
gi|30695777 [NCBI NR] gi|27311753 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 957 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
integral to membrane [IEA] |
| Biological Process: |
glucosylceramide catabolic process
[IEA]
sphingolipid metabolic process [IEA] |
| Molecular Function: |
glucosylceramidase activity
[IEA]
catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFEEKIMDIG EDVKPLNSSD TKVDPAVPAS LTWQRKIDSD VKAPREFNLS VKEIFQLAPV 60
61 GIRLWFLCRE EAAKGRLAFI DPFSKHSVTS SHGVPLGGIG AGSIGRSFKG EFQRWQLFPP 120
121 KCEDEPVLAN QFSAFVSRAN GKKYSSVLCP RNPKLDKQDS ESGIGSWDWN LKGDKSTYHA 180
181 LYPRSWTMYE GEPDPELRIV CRQVSPFIPH NYKESSFPVS VFTFTLHNLG NTTADVTLLF 240
241 TWANSVGGDS EFSGGHYNSK ITMNDGVQGV LLHHKTANGL PSLSYAISAQ ATDGVSVSAC 300
301 PFFIVSGKQD GITAKDMWQA VKENGSFDHL KASEASMQSD HGSSIGAAVA ASVTVLPGES 360
361 RIVTFSLAWD CPEVQFPSGK IYSRRYTKFY GNNGDAAAQI AHDAILGHSQ WESWIEDWQR 420
421 PILEDKRLPA WYPVTLFNEL YYLNSGGTLW TDGSSPVHSL AGVREKKFSL DKSQLGLKND 480
481 IDVPHQNDTA VSVLEKMAST LEELHASTTS NSAFGTKLLE EGEENIGHFL YLEGIEYRMW 540
541 NTYDVHFYAS FALVMLFPKL ELSIQRDFAA AVMLHDPTKV KTLSEGQWVQ RKVLGAVPHD 600
601 LGINDPWFEV NGYTLHNTDR WKDLNPKFVL QVYRDVVATG DKKFASAVWP SVYVAMAYMA 660
661 QFDKDGDGMI ENEGFPDQTY DTWSASGVSA YCGGLWVAAL QAASALARVV GDKNSQDYFW 720
721 SKFQKAKVVY EKKLWNGSYF NYDNSGSQYS STIQADQLAG QWYARASGLL PIVDEDKART 780
781 ALEKVYNYNV MKIKDGKRGA VNGMHPNGKV DTASMQSREI WSGVTYALSA TMIQEGLVEM 840
841 AFQTASGIYE AAWSETGLGY SFQTPESWNT VDEYRSLTYM RPLAIWAMQW ALTKTSQKQE 900
901 QLGLEPEQQE PELEPSSSMK HDIGFSRVSR LLSLPNEASA KSTLQTLFDY TCRRMMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
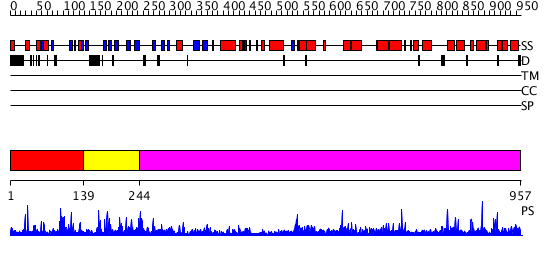
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 2.04098 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [139..243] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [244..957] | 65.30103 | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)