













| General Information: |
|
| Name(s) found: |
CE37708
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 226 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
arginine metabolic process
[IEA ectoine biosynthetic process [IEA tetrapyrrole biosynthetic process [IEA biosynthetic process [IEA biotin biosynthetic process [IEA gamma-aminobutyric acid metabolic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA 4-aminobutyrate transaminase activity [IEA transaminase activity [IEA glutamate-1-semialdehyde 2,1-aminomutase activity [IEA diaminobutyrate-pyruvate transaminase activity [IEA adenosylmethionine-8-amino-7-oxononanoate transaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSSLRRVVP SLPRGSRSLT SQQIFDREKK FGCHNYKPLP VALSKGEGCF VWDVEGKKYF 60
61 DFLAAYSAVN QGHCHPKLLK VVQEQASTLT LTSRAFYNNV LGEYEEYVTK LFKYDKVLPM 120
121 NTGVEACESA VKLARRWAYD VKGVKDNEAV VVFAENNFWG RSIAAISAST DPDSFARFGP 180
181 FVPGFKTVPY NNLKTSRTLP RQRPARYCCT RKSAFWRILP SFCSAL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
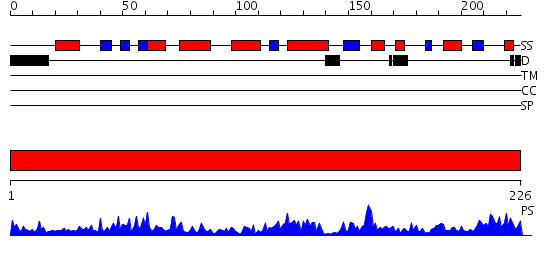
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..226] | 65.69897 | Ornithine aminotransferase mutant Y85I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)