













| General Information: |
|
| Name(s) found: |
INO80_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
positive regulation of DNA repair
[IMP]
regulation of transcription [IMP] somatic cell DNA recombination [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPSRRPPKD SPYANLFDLE PLMKFRIPKP EDEVDYYGSS SQDESRSTQG GVVANYSNGS 60
61 KSRMNASSKK RKRWTEAEDA EDDDDLYNQH VTEEHYRSML GEHVQKFKNR SKETQGNPPH 120
121 LMGFPVLKSN VGSYRGRKPG NDYHGRFYDM DNSPNFAADV TPHRRGSYHD RDITPKIAYE 180
181 PSYLDIGDGV IYKIPPSYDK LVASLNLPSF SDIHVEEFYL KGTLDLRSLA ELMASDKRSG 240
241 VRSRNGMGEP RPQYESLQAR MKALSPSNST PNFSLKVSEA AMNSAIPEGS AGSTARTILS 300
301 EGGVLQVHYV KILEKGDTYE IVKRSLPKKL KAKNDPAVIE KTERDKIRKA WINIVRRDIA 360
361 KHHRIFTTFH RKLSIDAKRF ADGCQREVRM KVGRSYKIPR TAPIRTRKIS RDMLLFWKRY 420
421 DKQMAEERKK QEKEAAEAFK REQEQRESKR QQQRLNFLIK QTELYSHFMQ NKTDSNPSEA 480
481 LPIGDENPID EVLPETSAAE PSEVEDPEEA ELKEKVLRAA QDAVSKQKQI TDAFDTEYMK 540
541 LRQTSEMEGP LNDISVSGSS NIDLHNPSTM PVTSTVQTPE LFKGTLKEYQ MKGLQWLVNC 600
601 YEQGLNGILA DEMGLGKTIQ AMAFLAHLAE EKNIWGPFLV VAPASVLNNW ADEISRFCPD 660
661 LKTLPYWGGL QERTILRKNI NPKRMYRRDA GFHILITSYQ LLVTDEKYFR RVKWQYMVLD 720
721 EAQAIKSSSS IRWKTLLSFN CRNRLLLTGT PIQNNMAELW ALLHFIMPML FDNHDQFNEW 780
781 FSKGIENHAE HGGTLNEHQL NRLHAILKPF MLRRVKKDVV SELTTKTEVT VHCKLSSRQQ 840
841 AFYQAIKNKI SLAELFDSNR GQFTDKKVLN LMNIVIQLRK VCNHPELFER NEGSSYLYFG 900
901 VTSNSLLPHP FGELEDVHYS GGQNPIIYKI PKLLHQEVLQ NSETFCSSVG RGISRESFLK 960
961 HFNIYSPEYI LKSIFPSDSG VDQVVSGSGA FGFSRLMDLS PSEVGYLALC SVAERLLFSI1020
1021 LRWERQFLDE LVNSLMESKD GDLSDNNIER VKTKAVTRML LMPSKVETNF QKRRLSTGPT1080
1081 RPSFEALVIS HQDRFLSSIK LLHSAYTYIP KARAPPVSIH CSDRNSAYRV TEELHQPWLK1140
1141 RLLIGFARTS EANGPRKPNS FPHPLIQEID SELPVVQPAL QLTHRIFGSC PPMQSFDPAK1200
1201 LLTDSGKLQT LDILLKRLRA GNHRVLLFAQ MTKMLNILED YMNYRKYKYL RLDGSSTIMD1260
1261 RRDMVRDFQH RSDIFVFLLS TRAGGLGINL TAADTVIFYE SDWNPTLDLQ AMDRAHRLGQ1320
1321 TKDVTVYRLI CKETVEEKIL HRASQKNTVQ QLVMTGGHVQ GDDFLGAADV VSLLMDDAEA1380
1381 AQLEQKFREL PLQVKDRQKK KTKRIRIDAE GDATLEELED VDRQDNGQEP LEEPEKPKSS1440
1441 NKKRRAASNP KARAPQKAKE EANGEDTPQR TKRVKRQTKS INESLEPVFS ASVTESNKGF1500
1501 DPSSSAN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
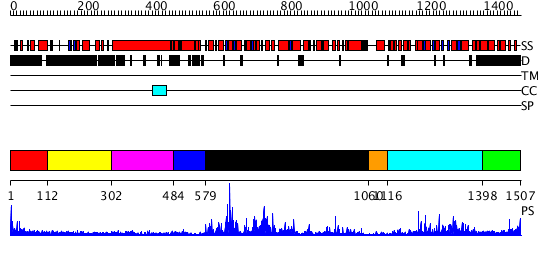
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 1.222993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [112..301] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [302..483] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [484..578] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [579..1059] | 86.0 | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | |
| 6 | View Details | [1060..1115] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1116..1397] | 41.30103 | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain | |
| 8 | View Details | [1398..1507] | 1.334996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)