













| General Information: |
|
| Name(s) found: |
gi|7446608
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Mycobacterium tuberculosis |
| Length: | 1289 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAGSRFPLV DPLPSVGARP DRLRGQPRRR TRAGGRPGSA RCVPEAAAAA AGRHDTGPRR 60
61 QSRRRLVAVD GADHRVQRAV IWPLVYLKSL TLKGFKSFAA PTTLRFEPGI TAVVGPNGSG 120
121 KSNVVDALAW VMGEQGAKTL RGGKMEDVIF AGTSSRAPLG RAEVTVSIDN SDNALPIEYT 180
181 EVSITRRMFR DGASEYEING SSCRLMDVQE LLSDSGIGRE MHVIVGQGKL EEILQSRPED 240
241 RRAFIEEAAG VLKHRKRKEK ALRKLDTMAA NLARLTDLTT ELRRQLKPLG RQAEAAQRAA 300
301 AIQADLRDAR LRLAADDLVS RRAEREAVFQ AEAAMRREHD EAAARLAVAS EELAAHESAV 360
361 AELSTRAESI QHTWFGLSAL AERVDATVRI ASERAHHLDI EPVAVSDTDP RKPEELEAEA 420
421 QQVAVAEQQL LAELDAARAR LDAARAELAD RERRAAEADR AHLAAVREEA DRREGLARLA 480
481 GQVETMRARV ESIDESVARL SERIEDAAMR AQQTRAEFET VQGRIGELDQ GEVGLDEHHE 540
541 RTVAALRLAD ERVAELQSAE RAAERQVASL RARIDALAVG LQRKDGAAWL AHNRSGAGLF 600
601 GSIAQLVKVR SGYEAALAAA LGPAADALAV DGLTAAGSAV SALKQADGGR AVLVLSDWPA 660
661 PQAPQSASGE MLPSGAQWAL DLVESPPQLV GAMIAMLSGV AVVNDLTEAM GLVEIRPELR 720
721 AVTVDGDLVG AGWVSGGSDR KLSTLEVTSE IDKARSELAA AEALAAQLNA ALAGALTEQS 780
781 ARQDAAEQAL AALNESDTAI SAMYEQLGRL GQEARAAEEE WNRLLQQRTE QEAVRTQTLD 840
841 DVIQLETQLR KAQETQRVQV AQPIDRQAIS AAADRARGVE VEARLAVRTA EERANAVRGR 900
901 ADSLRRAAAA EREARVRAQQ ARAARLHAAA VAAAVADCGR LLAGRLHRAV DGASQLRDAS 960
961 AAQRQQRLAA MAAVRDEVNT LSARVGELTD SLHRDELANA QAALRIEQLE QMVLEQFGMA1020
1021 PADLITEYGP HVALPPTELE MAEFEQARER GEQVIAPAPM PFDRVTQERR AKRAERALAE1080
1081 LGRVNPLALE EFAALEERYN FLSTQLEDVK AARKDLLGVV ADVDARILQV FNDAFVDVER1140
1141 EFRGVFTALF PGGEGRLRLT EPDDMLTTGI EVEARPPGKK ITRLSLLSGG EKALTAVAML1200
1201 VAIFRARPSP FYIMDEVEAA LDDVNLRRLL SLFEQLREQS QIIIITHQKP TMEVADALYG1260
1261 VTMQNDGITA VISQRMRGQQ VDQLVTNSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
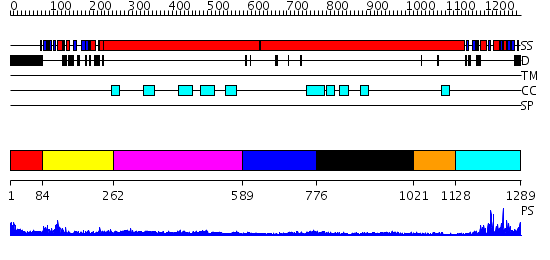
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..261] | 42.0 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 3 | View Details | [262..588] | 3.2 | Tropomyosin | |
| 4 | View Details | [589..775] | 4.9 | Smc hinge domain | |
| 5 | View Details | [776..1020] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1021..1127] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1128..1289] | 20.154902 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
|||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)