













| General Information: |
|
| Name(s) found: |
DNAJ_PSEAE
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Pseudomonas aeruginosa PAO1 |
| Length: | 377 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRDFYEVL GVERGASEAD LKKAYRRLAM KYHPDRNPGD KEAEDKFKEA NEAYEVLSDA 60
61 SKRAAYDQYG HAGVDPNMGG GAGAGFGGAS FSDIFGDVFS DFFGGGGARG GSRGGAQRGA 120
121 DLRYTLDLDL EEAVRGTTVT IRVPTLVGCK TCNGSGAKPG TTPVTCTTCG GIGQVRMQQG 180
181 FFSVQQTCPR CHGTGKMISD PCGSCHGQGR VEEQKTLSVK VPAGVDTGDR IRLTGEGEAG 240
241 SMGGPAGDLY VVVNVREHPI FQRDGKHLYC EVPISFADAA LGGELEVPTL DGRVKLKIPE 300
301 STQTGKLFRL RGKGVAPVRG GGAGDLMCKV VVETPVNLDK RQRELLEEFR KSLQSDTSHS 360
361 PKASGWFEGM KRFFDDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
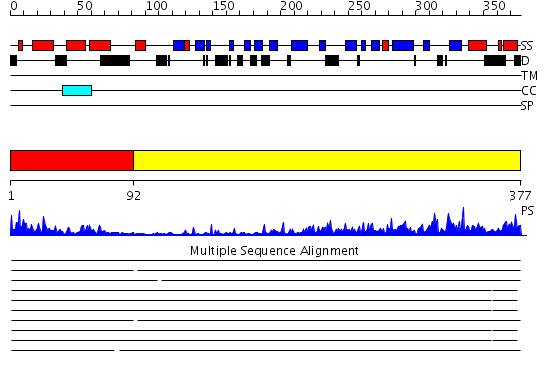
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 20.522879 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 2 | View Details | [92..377] | 42.09691 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)