













| General Information: |
|
| Name(s) found: |
PIE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2055 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[ISS]
cell wall [IDA] chromatin remodeling complex [IDA] |
| Biological Process: |
petal development
[IGI]
petal formation [IGI] response to cadmium ion [IDA] defense response to bacterium [IGI] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASKGGKSKP DIVMASKSGK SKPDNESRAK RQKTLEAPKE PRRPKTHWDH VLEEMAWLSK 60
61 DFESERKWKL AQAKKVALRA SKGMLDQASR EERKLKEEEQ RLRKVALNIS KDMKKFWMKV 120
121 EKLVLYKHQL VRNEKKKKAM DKQLEFLLGQ TERYSTMLAE NLVEPYKQGQ NTPSKPLLTI 180
181 ESKSDEERAE QIPPEINSSA GLESGSPELD EDYDLKSEDE TEDDEDTIEE DEKHFTKRER 240
241 QEELEALQNE VDLPVEELLR RYTSGRVSRE TSPVKDENED NLTSVSRVTS PVKDENQDNL 300
301 ASVGQDHGED KNNLAASEET EGNPSVRRSN DSYGHLAISE THSHDLEPGM TTASVKSRKE 360
361 DHTYDFNDEQ EDVDFVLANG EEKDDEATLA VEEELAKADN EDHVEEIALL QKESEMPIEV 420
421 LLARYKEDFG GKDISEDESE SSFAVSEDSI VDSDENRQQA DLDDDNVDLT ECKLDPEPCS 480
481 ENVEGTFHEV AEDNDKDSSD KIADAAAAAR SAQPTGFTYS TTKVRTKLPF LLKHSLREYQ 540
541 HIGLDWLVTM YEKKLNGILA DEMGLGKTIM TIALLAHLAC DKGIWGPHLI VVPTSVMLNW 600
601 ETEFLKWCPA FKILTYFGSA KERKLKRQGW MKLNSFHVCI TTYRLVIQDS KMFKRKKWKY 660
661 LILDEAHLIK NWKSQRWQTL LNFNSKRRIL LTGTPLQNDL MELWSLMHFL MPHVFQSHQE 720
721 FKDWFCNPIA GMVEGQEKIN KEVIDRLHNV LRPFLLRRLK RDVEKQLPSK HEHVIFCRLS 780
781 KRQRNLYEDF IASTETQATL TSGSFFGMIS IIMQLRKVCN HPDLFEGRPI VSSFDMAGID 840
841 VQLSSTICSL LLESPFSKVD LEALGFLFTH LDFSMTSWEG DEIKAISTPS ELIKQRVNLK 900
901 DDLEAIPLSP KNRKNLQGTN IFEEIRKAVF EERIQESKDR AAAIAWWNSL RCQRKPTYST 960
961 SLRTLLTIKG PLDDLKANCS SYMYSSILAD IVLSPIERFQ KMIELVEAFT FAIPAARVPS1020
1021 PTCWCSKSDS PVFLSPSYKE KVTDLLSPLL SPIRPAIVRR QVYFPDRRLI QFDCGKLQEL1080
1081 AMLLRKLKFG GHRALIFTQM TKMLDVLEAF INLYGYTYMR LDGSTPPEER QTLMQRFNTN1140
1141 PKIFLFILST RSGGVGINLV GADTVIFYDS DWNPAMDQQA QDRCHRIGQT REVHIYRLIS1200
1201 ESTIEENILK KANQKRVLDN LVIQNGEYNT EFFKKLDPME LFSGHKALTT KDEKETSKHC1260
1261 GADIPLSNAD VEAALKQAED EADYMALKRV EQEEAVDNQE FTEEPVERPE DDELVNEDDI1320
1321 KADEPADQGL VAAGPAKEEM SLLHSDIRDE RAVITTSSQE DDTDVLDDVK QMAAAAADAG1380
1381 QAISSFENQL RPIDRYAIRF LELWDPIIVE AAMENEAGFE EKEWELDHIE KYKEEMEAEI1440
1441 DDGEEPLVYE KWDADFATEA YRQQVEVLAQ HQLMEDLENE AREREAAEVA EMVLTQNESA1500
1501 HVLKPKKKKK AKKAKYKSLK KGSLAAESKH VKSVVKIEDS TDDDNEEFGY VSSSDSDMVT1560
1561 PLSRMHMKGK KRDLIVDTDE EKTSKKKAKK HKKSLPNSDI KYKQTSALLD ELEPSKPSDS1620
1621 MVVDNELKLT NRGKTVGKKF ITSMPIKRVL MIKPEKLKKG NLWSRDCVPS PDSWLPQEDA1680
1681 ILCAMVHEYG PNWNFVSGTL YGMTAGGAYR GRYRHPAYCC ERYRELIQRH ILSASDSAVN1740
1741 EKNLNTGSGK ALLKVTEENI RTLLNVAAEQ PDTEMLLQKH FSCLLSSIWR TSTRTGNDQM1800
1801 LSLNSPIFNR QFMGSVNHTQ DLARKPWQGM KVTSLSRKLL ESALQDSGPS QPDNTISRSR1860
1861 LQETQPINKL GLELTLEFPR GNDDSLNQFP PMISLSIDGS DSLNYVNEPP GEDVLKGSRV1920
1921 AAENRYRNAA NACIEDSFGW ASNTFPANDL KSRTGTKAQS LGKHKLSASD SAKSTKSKHR1980
1981 KLLAEQLEGA WVRPNDPNLK FDFTPGDREE EEEQEVDEKA NSAEIEMISC SQWYDPFFTS2040
2041 GLDDCSLASD ISEIE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
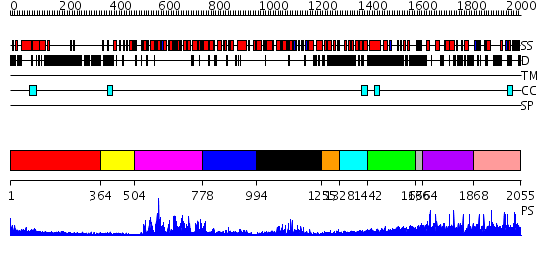
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..363] | 19.39794 | Heavy meromyosin subfragment | |
| 2 | View Details | [364..503] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [504..777] | 88.09691 | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | |
| 4 | View Details | [778..993] | 5.08 | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain | |
| 5 | View Details | [994..1254] | 25.69897 | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain | |
| 6 | View Details | [1255..1327] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1328..1441] | 7.221849 | nucleosome recognition module of ISWI ATPase | |
| 8 | View Details | [1442..1635] | 4.69897 | No description for 2dfsA was found. | |
| 9 | View Details | [1636..1663] | 5.522879 | No description for 2i1jA was found. | |
| 10 | View Details | [1664..1867] | 2.44 | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | |
| 11 | View Details | [1868..2055] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. | ||||||
| 9 | No functions predicted. | ||||||
| 10 |
|
||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)