













| General Information: |
|
| Name(s) found: |
HGNC:24358|MIM:300564|Ensembl:ENSG00000184205|HPRD:06713,HsxXG006207,C06,HsCD00301930,470807.0,pANT7_cGST,FUSION,AL713652,64061.0,TSPYL2,CDA1|CINAP|CTCL|DENTT|HRIHFB2216|NP79|SE204|TSPX,TSPY-like
[120926-122011-mus-fusion.fasta]
|
| Description(s) found: |
|
| Organism: | SPECIES UNKNOWN |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRPDEGPPA KTRRLSSSES PQRDPPPPPP PPPLLRLPLP PPQQRPRLQE ETEAAQVLAD 60
61 MRGVGLGPAL PPPPPYVILE EGGIRAYFTL GAECPGWDST IESGYGEAPP PTESLEALPT 120
121 PEASGGSLEI DFQVVQSSSF GGEGALETCS AVGWAPQRLV DPKSKEEAII IVEDEDEDER 180
181 ESMRSSRRRR RRRRRKQRKV KRESRERNAE RMESILQALE DIQLDLEAVN IKAGKAFLRL 240
241 KRKFIQMRRP FLERRDLIIQ HIPGFWVKAF LNHPRISILI NRRDEDIFRY LTNLQVQDLR 300
301 HISMGYKMKL YFQTNPYFTN MVIVKEFQRN RSGRLVSHST PIRWHRGQEP QARRHGNQDA 360
361 SHSFFSWFSN HSLPEADRIA EIIKNDLWVN PLRYYLRERG SRIKRKKQEM KKRKTRGRCE 420
421 VVIMEDAPDY YAVEDIFSEI SDIDETIHDI KISDFMETTD YFETTDNEIT DINENICDSE 480
481 NPDHNEVPNN ETTDNNESAD DHETTDNNES ADDNNENPED NNKNTDDNEE NPNNNENTYG 540
541 NNFFKGGFWG SHGNNQDSSD SDNEADEASD DEDNDGNEGD NEGSDDDGNE GDNEGSDDDD 600
601 RDIEYYEKVI EDFDKDQADY EDVIEIISDE SVEEEGIEEG IQQDEDIYEE GNYEEEGSED 660
661 VWEEGEDSDD SDLEDVLQVP NGWANPGKRG KTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
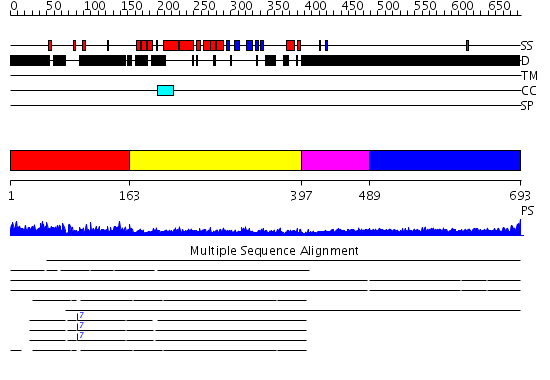
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [163..396] | 29.0 | No description for 2e50A was found. | |
| 3 | View Details | [397..488] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [489..693] | 1.469995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)