













| General Information: |
|
| Name(s) found: |
gi|24376373
[NCBI NR]
gi|24376247 [NCBI NR] gi|24345287 [NCBI NR] gi|24344713 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 988 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
transposition, DNA-mediated
[ISS |
| Molecular Function: |
transposase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRRSILSAA ERDSLLVLPD TQDELIRHYT FSEPDLSLIR QRRGDANRLG IAVQLCLLRF 60
61 PGQGLLPDAT VPMPLLQWIG QQLQLDPVCW PQYAEREETR REHLLELRVY LGMEPFSQVH 120
121 HRQAVHTTTE LALQTDKGIV LANSVVETLR HKHIILPTLD VVERVCAEAL TRANRRIYDT 180
181 LTEPLSGSHR HRLDDLLKLR DNNKTTTLAW LRLSPVKPNS RHMLEHIERL KVWQALDLPI 240
241 GVDRLIHQNR LLKIAREGGQ MTPADLAKFE PQRRYATLVA LAIEGMATVT DEIIDLHDRI 300
301 MGKLFNDAKK RHQKQFQASG KAINAKVRLF GRIGQVLIDA KQAGDDPFAA IEGVISWEAF 360
361 AKSVTEAQSL AQPEEFDFLY RLGESYATLR RYAPTFLTAL KLRAAPAAKG VLEAIEVLRS 420
421 MNNDNARKVP ADAPIDFIKP RWQKLVITDT GIDRRYYELC ALSEMRNALR SGDIWVQGSR 480
481 QFKDFEDYLV PPAKFVSLKQ TNQLPLAVAT DCEQYLNERL TQLETQLATV NSMAQANELP 540
541 DAIITASGLK ITPLDAVVPD TAQRLIDQAA RILPHVKITE LLLEVDEWTG FTRHFAHLKS 600
601 GVLAKDKNLL LTTILADAIN LGLTKMAESC PGTTYAKLAW LQAWHIRDET YGAALSELVN 660
661 AQYRHPFAEH WGDGSTSSSD GQNFRTGNKA ESTGHINPKY GSSPGRTFYT HISDQYAPFH 720
721 TKVVNVGVRD STYVLDGLLY HESDLRIEEH YTDTAGFTDH VFALMHLLGF RFAPRIRDLG 780
781 ETKLYIPKRD VTYEGLKSMI GGTLNIKLIR THWDEILRLA TSIKQGTVTA SLMLRKLGSY 840
841 PRQNGLAVAL RELGRIERTL FILDWLQSVE LRRRVHAGLN KGEARNALAR AVFFNRLGEI 900
901 RDRSFEQQRY RASGLNLVTA AIVLWNTVYL ERVAHGLRAK GHAVDEELLQ YLSPLGWEHI 960
961 NLTGDYLWRS SAKIGSGKFR PLRPLSPA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
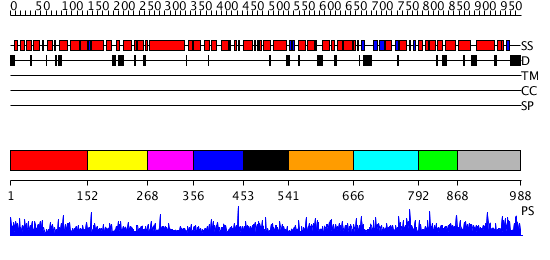
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 9.078994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [152..267] | 3.073985 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [268..355] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [356..452] | 1.073991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [453..540] | 1.075994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [541..665] | 2.080963 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [666..791] | 3.091983 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [792..867] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [868..988] | 13.107978 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)