













| General Information: |
|
| Name(s) found: |
gi|24375542
[NCBI NR]
gi|24350421 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 797 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
methionine biosynthetic process
[ISS lysine biosynthetic process via diaminopimelate [ISS threonine biosynthetic process [ISS |
| Molecular Function: |
aspartate kinase activity
[ISS homoserine dehydrogenase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARCHLHKFG GSSLADADCY RRVAHILLTH GHSDDLVVVS AAGKTTNFLY KLLSLRDSNQ 60
61 LWQEELQVLI SYQQGLIEQL LSNEQARDLR ERLTTDKAQL ISLLSLDVRN DYQISHVVSF 120
121 GERWSARLMA ALLRESGVAA SHVDACSILV ADEAAVPQIR VQESRAKVQA LLAAHPNERL 180
181 VITGFICANE RGDTLLLGRN GSDFSASLIA SLADIERVTI WTDVEGVFNA DPNKINDAKL 240
241 LKSMSLAEAD RLARLGSPVL HSRTLQPLFN TEVSLAVRSS YASHTDFTLI APHSSSASAP 300
301 VVTNLNAVVL FGFKIAGDVA SLLDQLVEAG LTPLAHWVSA HQRLELAYTA ENQKQVQKLL 360
361 DAQAEALGIR DLQINTELGL VALVSADAEH YRRSFARLLS RDAKPLFSGD LSLVTLVPQS 420
421 QVNLLTQKVH RRCAGPRKRI GVLLLGVGNI GEAWVKLFKS VSPSLNQELE AQVELVGLVT 480
481 SSKALIKSKG VDLQSWQQEF DNEATPWQYE HLFEQLEQLN CDELIALDIS ASASLTLQYP 540
541 EFFERGIHMV SANKLAGSGP LPFYRELKEQ LGYRRLFWRY NASCGAGLPI QHALNDLRNS 600
601 GDSVEAVGGI FSGTLCWLFE KYDTSKPFSE LVIEARGLGI TEPDPRDDLS GRDMQRKLLI 660
661 LAREIGLEIE LEDIELRSLV PAHLADIPLE QFLARIGELD DELLQQYGAA AEQNKVLRYV 720
721 ASLDNSGAVL KAEVGLQWID ASHPYANLTP GDNVFVIRSA FYQGNPLIIR GPGAGREVTA 780
781 AAVQSDLTQI CRDLLQE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
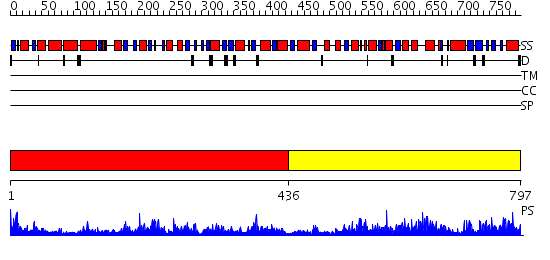
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..435] | 88.0 | No description for 2hmfA was found. | |
| 2 | View Details | [436..797] | 64.522879 | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)