













| General Information: |
|
| Name(s) found: |
gi|24373032
[NCBI NR]
gi|24347201 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Type I site-specific deoxyribonuclease complex
[ISS |
| Biological Process: |
DNA modification
[ISS |
| Molecular Function: |
DNA-methyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTDNDKKQQ EVLAIEDGMM LDYLTNNPIK ETPKELVRQK TLRALFHEYG LSAEDMALDF 60
61 NTTVGGKRKK IDIAIFEHGA EHLSENIRRI VICDKEPIKG KKSAVKMRDH DQAQKDLQLM 120
121 EDMMREQVGD KYVLEKCHWG LWTNGIEFFF VEKEESRFDT KFNAVGDWPL ADETLGSRDV 180
181 ASNAQLRTAD REMLLTAFRR CHNFIHGNEG MPKDAAFWQF LYLIFSKMYD ERIGNRDREF 240
241 WASPTEQFDD EGRKKIRARI NPLFEKVKKA YPEIFSGNEE IILSDRALAF MVSELAKYDF 300
301 TRTEMDAKGA AYQEVVGDNL RGDRGQYFTP RGAIKLIVEM MAPQPHEKVL DPSCGTGGFL 360
361 EQTLSFINRK LCEEEEVKLG AETTEEFISI QQQIKKFAEN NLFGCDFDPF LCRASQMNVV 420
421 MASNAMANIY HMNSLEYPHG HLKGVEPAKS KIPVGDSSGK DGSIDVILTN PPFGSDIPVT 480
481 DKQILEQYDL AYVWERTENG GFRKTERRKD AVSPEILFIE RCVQWLKQGG RMGIVLPDGI 540
541 LGNPGDEYIR WWLMQECWVL GCVDLPVESF IVEANVNILT SLLFLKKKTD TEKDAIAFGG 600
601 EPEYPVFMAV AEKVGFDRRG NTLYERHPDG EEKVIEEDVE ERIRINGQNV VRKLKRRSKI 660
661 LDDDLPKIAK AWKEFRANNP EPSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
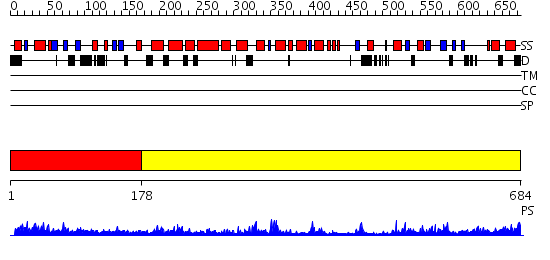
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | 7.314995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [178..684] | 55.69897 | Crystal structure of Type I restriction enzyme EcoKI M protein (EC 2.1.1.72) (M.EcoKI) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)