













| General Information: |
|
| Name(s) found: |
gi|24372945
[NCBI NR]
gi|24347085 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
L-phenylalanine biosynthetic process
[ISS |
| Molecular Function: |
chorismate mutase activity
[ISS prephenate dehydratase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKPQPLNHT REQITQLDND LLSLLAERRR LSLEVARSKE VDVRPIRDTQ REKELLARLV 60
61 TAGREKGLDA HYVISLYQSI IEDSVLNQQA YLHGRANPET QKQQYCIAYL GARGSYSYLA 120
121 ASRYCQRRQV EMLDLGCQSF DEIVQAVESG HADYGFLPIE NTSSGSINEV YDVLQHTSLS 180
181 IVGETTIEVS HCLLGKPGSK LSEIKTVYAH PQPISQCSRY LSQHKALRLE YCSSSAEAME 240
241 KVNQSPDNSA AAIGSAEGGA LYQLESIESG LANQKINQSR FIVVARKAVA VPEQLPAKTT 300
301 LIMATGQKAG ALVEALLVLK AHQLNMSKLE SRPIPGTPWE EMFYLDIDAN ISSEAMQQGL 360
361 KQLERITRFI KVLGCYPCET VTPTQLSNSQ LLIEPNTSKA EVLSEVSTKQ SPYRYSKAYK 420
421 AQASEIHCGP FTIGAGHIGA IAKVTLSKSL LHLESSHAAA HIALSAFEHK IKQLKEVGFQ 480
481 AVILEGCQQL ASKEAIIPKL RQTLHQYDLL CVIAIEQAAD MPLATEHADM LFLTGKQMFN 540
541 QALLTQAGTL PIPLFLERND MASYEEFLAA TETILSQGNQ QLILCDSGIR TYNNANLPTL 600
601 DLASLILIKA NSHLPIVINP CYAINEDALV LQTQGIKQLK ADGIVLNYAL EEDKAHHSLA 660
661 LMGEVVRELY R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
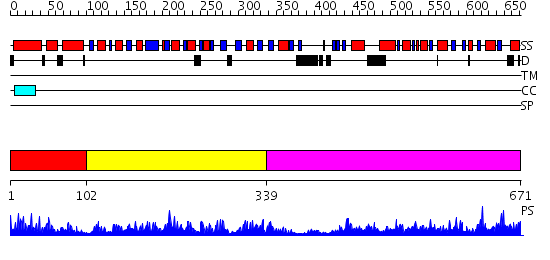
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 20.69897 | Chorismate mutase domain of P-protein | |
| 2 | View Details | [102..338] | 78.09691 | No description for 2qmxA was found. | |
| 3 | View Details | [339..671] | 74.522879 | Crystal structure of Phospho-2-dehydro-3-deoxyheptonate aldolase (DAHP synthase) (TM0343) from Thermotoga Maritima at 1.92 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)