













| General Information: |
|
| Name(s) found: |
gi|24372910
[NCBI NR]
gi|24347039 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 744 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
phosphoenolpyruvate-dependent sugar phosphotransferase system
[ISS |
| Molecular Function: |
protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNTLRDITQ AVASAHSLET ALEVLVSSTK AAMETQCCSV YILEQQELVL SATDGLEKSA 60
61 VGRVRMPLTQ GLVGLAAERE EAVNLADARL HPRFKLFPEV AEEEYRAFLA VPIIYQKAVV 120
121 GVIVVQQASA RQFSEGEEAF LMTLAAQLAM AIRGLKQKAQ VSSLHQQILF QGTSASSGIA 180
181 IAHAFVLGGE ISLEQPDIRC EDIVLESNRL VAAMGRCKEA IGALSQRFDR EQDEEVASIF 240
241 NALQLLLDDA SLGGEYAREV LLGWEAESAV SRVSLRYIQQ FLAMEDPYLK ERASDIRDLG 300
301 QKVLRQLIEP ERLELEPDKP VILVTKEADA TMLAEFPRQK LAGIVTELGG VNSHAAILAR 360
361 ALGVPAITGV EQLLSADIDQ KLLVVNASRG QLMVSPSPAI VSEYRSLISA QKALQRQYAQ 420
421 ELTLPSVMLD GKRIRLYLNA GLLSGVASEI AEGADGIGLY RTEIPFMLQQ RFPSESEQVK 480
481 VYQQVLSAAS GRPVVMRTLD VGGDKPLPYF PIKEDNPFLG WRGIRLSLDH PELFLVQLRA 540
541 MLQAGAEGKQ LSILLPMVSN LDEIDQALAY LEQAHVELKN DVNSNIKMPR IGIMLEVPAM 600
601 LYQLDEVAKR VDFVSVGSND LTQYLLAVDR NNPRVSSLFD SYHPGILRAL HQARLDCEYH 660
661 HLDISICGEL AGEPMGAILL VAMGYHQLSM NQGSLARINY LLRRVSGEDL AQLLAQALSL 720
721 SNGFQVRELV KEYLTLQGLV TILN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
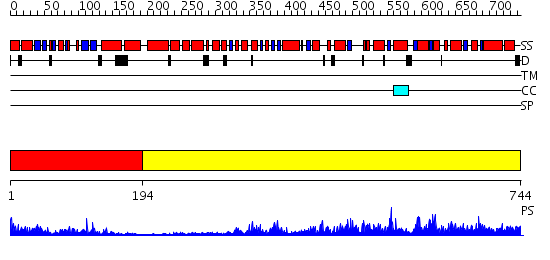
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 19.0 | 3',5'-cyclic nucleotide phosphodiesterase 2A, GAF A and GAF B domains | |
| 2 | View Details | [194..744] | 149.0 | No description for 2hroA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)