













| General Information: |
|
| Name(s) found: |
SYGB_SHEON
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 688 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycyl-tRNA aminoacylation
[ISS |
| Molecular Function: |
glycine-tRNA ligase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFENLLIEL GTEELPPKAL RKLAESFLAN FTEELTKADL AFKSAVWYAA PRRLALNITE 60
61 LAIAQADKIV EKRGPAVSSA FDAGGKPTKA AEGWARGNGI TVDQAERLVT DKGEWLVYNA 120
121 KVEGVETKSL VAAMAQRALD KLPIPKPMRW GSSKTQFIRP VHTATMLLGS ELIEGELLGI 180
181 KSARNIRGHR FMGTGFELDH ADNYLTLLKE KGKVIADYES RKALIKADAE KAAAKIGGTA 240
241 DIEDALLEEV TSLVEWPVVL TASFEEKFLS VPSEALVYTM KGDQKYFPVF DDAGKLLPNF 300
301 IFVANIESKD PAQIISGNEK VVRPRLADAE FFFNTDKKHT LESRLPSLET VLFQQQLGTL 360
361 KDKVTRISAL AAFIAEQTGA NAVDAARAGL LSKTDLMTNM VMEFTDTQGT MGMHYARLDG 420
421 ETEAVALAME EQYKPKFSGD TVPTAAVSCA VALADKLDTL VGIFGIGQAP KGAADPFALR 480
481 RAAIGVLRII VENKLPLDLV TLIAKAQELH GANLSNANAS EEVLEFLMAR FRAWYQDKGI 540
541 NVDVILAVLA RRPTRPADFD SRINAVSHFR GLEASSALAA ANKRVSNILA KVEGELPSSV 600
601 NVALLSEAAE QALAAKLAEL QPQLAPLFAN ADYQQALTLL ASLRESVDQF FEDVMVMADD 660
661 AALRNNRLAL LNNLREQFLH VADISLLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
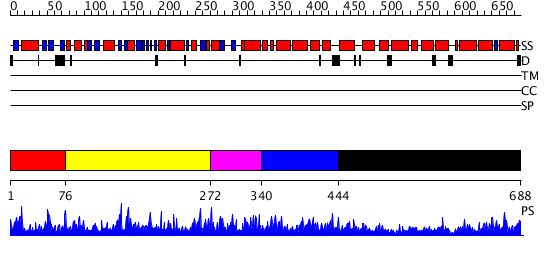
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 3.068989 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [76..271] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [272..339] | 1.071983 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [340..443] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [444..688] | 40.221849 | Arginyl-tRNA synthetase (ArgRS); Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)