













| General Information: |
|
| Name(s) found: |
YPP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNSNVRIPP TVPSKIIDVV DQALRARLLG GSTFNSGFDS LDSVLNLQFR LHYHVIGSNG 60
61 PAKPVCDVLL KESQNLEKNM SMMEELNDYP EITKLVEKIL FNCLGILFFH RGQFQESQRC 120
121 LLHSLKIHNN TASQKTALME QYDRYLIVEN LYYRGLVSQD INIMQNVFYK ELLAHVDTIP 180
181 PESNGLLFEY ISLIVAKLRF NQIQDLAENF KTTVENPFIL FLYMIKKFQS PLKKHIDNDD 240
241 LYLKFGQNVL LKAKFPTASE TNDEALEHFN VFLQYYFKFT HIKKIKVNPS WYNFIISSME 300
301 KTFQSIEVSK TAMFLFQNLS DNSNDEIKKK TFKRESILNF VNFVKYNDKY YQLHDNSHRD 360
361 IISFIDAYSF ILQNSSKTDS IENVFDYDNT VSTFATSLNS FYKEYNLPLM SQSESLDWLE 420
421 NSTRCVYPGN ISKVLTNAWS TLYEIRKYQL DFLVSNNLTS YLCNAMMLST KEKDNADVEE 480
481 QEEGEEEKAL RELQFKYSYT LAQQRHIETA IKTLESLILS KNPNYYKAWH LLALCRSVQE 540
541 DKEMSYKIVC SVLEAMNESL QNNTLLLNDR WQFIHLKLTQ LALIEEIFGT LEALETLPEV 600
601 FELYATLFPD SQPELNSMGP KYSQTKEYLL QMVWIFAANM YMRTKDNDED AKAAIKEASN 660
661 VESKFKNLNC NIANGYLSII KDEPGVALKE FETVLYYDEN NLDALVGFAE LIFPEELGVE 720
721 ETNLERYYTL SLDKKPGKRA KLTFVNDTDR SAAYARLKFL LECAILESIE AYYSPEVWWY 780
781 LSLIYEKYQD DEYKNSLLKC IKYQELNPIR SLRYCNY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
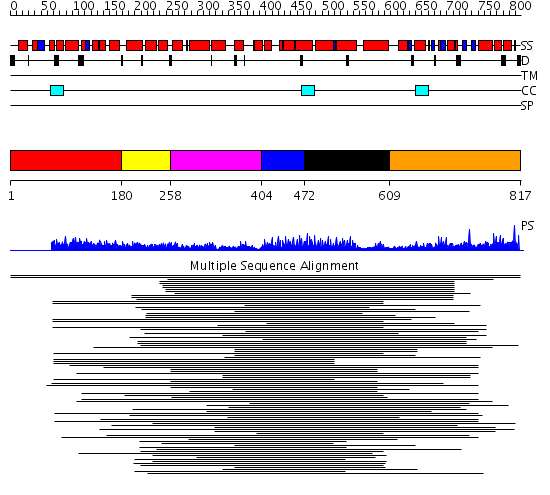
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 15.221849 | Hop | |
| 2 | View Details | [180..257] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [258..403] | 30.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [404..471] | 30.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [472..608] | 30.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [609..817] | 1.657995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)