













| General Information: |
|
| Name(s) found: |
YP216_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1102 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLLNRRKIQ PKEIGQSADS FSETPWVIKE SSERINDYDS DLKKLDFYKR DIFTCEISGK 60
61 DGLSYFKALK SEEQHREKVR YLLPKELRKA IANFANFSPI RKVGHLVESA FQRFSNRFFI 120
121 GDTVCLKTIQ KNALITYKEG EPNLVESPTI ENNVTLFLVK DVFQSNGMME SEKGEISAPK 180
181 LSLYLITECL NRESKGAALI VGQNEIKRPE SHFSKFIIAC FLNEILIKVS NKEHAPWRVK 240
241 QEYIERYDVN PKCSPNMIDY LPDRMNSSSS ELYTPLTIPP ESDVEPADWK ETSETSETSE 300
301 TSLSKIKAID DEISVSFDHI YDNVNSLAYN DLKGTVDDKE LPFTGPSIPF ENISYLDSSL 360
361 EYKNIDQKWF KECSQFPTER LLVVYQFLSF FGRFIGLSHF NFDQFLTTIK CTSPEALVDE 420
421 YVKINFLKTY NSKGSFTNEK PRNEIYNQVT SSNVSQREKA NVFNADESQR IPSNFTRNQK 480
481 MRKFITDKST EFVMYSIFKG KPLKNDDMEF QSYEKVNILY IDIVCSLMCL MTDNEPDWNC 540
541 NLMDNWTEEK RKEEGNKTEI DIAIEKCLNY GDTSWVKLLH NKNFSNGNWL ICLLGILQQN 600
601 THMIAYSDVA KCITKKILPL SMNFVNLGDE LWDNFRKRLS IKDKIDVLWV LVDFASNFSS 660
661 YIKELVDKVP KLCNGIRLKL DSAKKEYIKL KRQLKTLTKN RVKLHSNVSM NRYGSDECKG 720
721 KVNALKVKIA YLMEDIAFLE AKLIQSDIKR LEILGKDRNG NRYYWMDSNG SSSAISEKNE 780
781 ELYNCCFLWV QGPSEADINF CLDVDVESLK KWELLAKAKG TAYATKEVFS IFRSTDGSYY 840
841 QIAQGENFMI INSNGILMRP TIPAFIDKKI ISETPEKLLL SHHQWAFFND IEDIHMLVDR 900
901 LDDLRENEGQ LKKALTSKMD RIEVSYKQQF KIKRRIECDE TFKKNHKLLK NNEFTFPELK 960
961 RIETTCTSNG QHFSNMEKIS KKLSRTKNDL VLEAILKDVA HLGECERALL KKQQNLIYPL1020
1021 NFHFEQLRTI DLEFIVETKR KRQEDILTKL LNHQRYKHIS HVSGYGISSQ RVDKAAHLDV1080
1081 QGILEEIECQ LISRRREDEE RN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
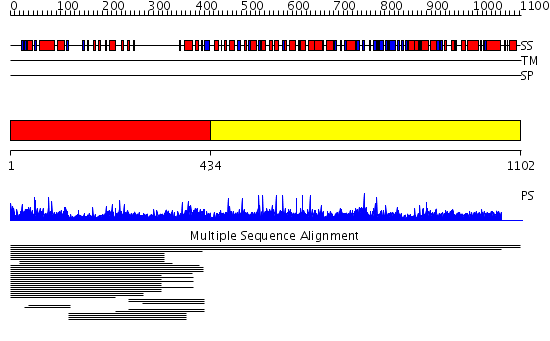
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..433] | 6.017933 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [434..1102] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..432] | 3.046996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [433..585] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [586..678] | 1.11 | Dimerization motif of sir4 | |
| 6 | View Details | [679..1102] | 1.67 | No description for 2oevA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)