













| General Information: |
|
| Name(s) found: |
YOR22_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRFTHRGLP SSTRFRNIFV RLNHIYVPWF YAIDVPNSKP YLPTYQTLHS PKKFKPFSVD 60
61 DSNRLEKASK RQERRPVLVN EDYLFKVDLS HMELSPTYWE GPTYQVRRGV WFDSSNQPLS 120
121 SDLTSEIEGL YKQLKFDDSN DDPTTTPPAE SQDIFRLKGK YPVDKENEGE QKNGSSNKDE 180
181 NESTFKFILF ANKQTAFLLS DLDGGKLQLA FLRSNLAQSL PINATMITRS YKYSSSATTK 240
241 QTSTSFKAAK TPQTEVADGS NSSKSRSIET KLEKKVSNLF NLSDFLQLFN GNASKDQDDA 300
301 QSLEKQMETD YNNADNSQGA NASSKIEDGK NSGASDRQIR SNRRDVDNLI LCVHGIGQTL 360
361 GKKYEYVNFA HTVNLLRSNM KKIYNNSEKL QSLNTAPDYK SNCNVQVLPI TWRHSISFQT 420
421 DAKEENIENP DLPTLSQVTV NGVLPLRKLL ADGLLDILLY VEPYYQDMIL QQVTSQLNKT 480
481 YRIFKEFNPE FDGKVHLVGH SLGSMILFDI LSKQKKYELE FQVDNLFFIG SPIGLLKLIQ 540
541 RTKIGDRPEF PNDLERKLTV QRPQCKDIYN VYHVCDPISY RMEPLVSKEM AHYEQTYLPH 600
601 CSEAYGLTSK VLEFGENIWK DLPGTDENNL QSKKTSPEKK EVKLSENLTR MLTGLNYTGR 660
661 LDYAMSPSLL EVDFISAIKS HVSYFEEPDI AAFILKEILS KHENASEIYV KRKTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
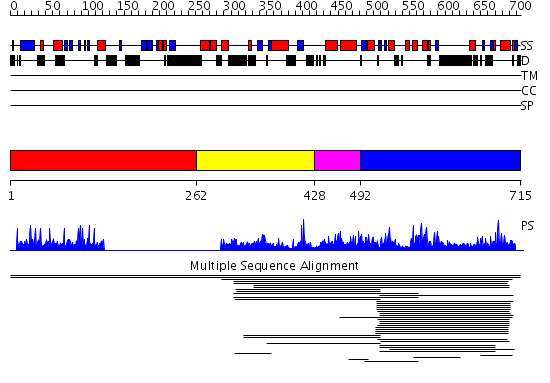
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [262..427] | 1.011988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [428..491] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [492..715] | 89.568636 | DDHD domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)