













| General Information: |
|
| Name(s) found: |
YL278_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1341 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRPRKNVSQ EKIQQLKREL ELAGNRTDVL LQDKKGRSRS CLLCRRRKQR CDHKLPSCTA 60
61 CLKAGIKCVQ PSKYSSSTSN SNTNNNTPTA GTVPPTPHPV IKRELQDSSI GAGAGAATSL 120
121 NDMTIIKPIS TSNSNVDAGD ANEFRKTIKS VTTNSNPNLM RQDKDQYTIF LEKKLKSLET 180
181 LLDLSPGCNQ YNYELSQYKK VSHLFSNNTS DYSRPNSSNM VILPLPSPSN KPLENTNNNG 240
241 SNVNAATNDT SASTNNINNN NAICQSASLL NDPLETLDFT KCIFAKYNLK KEFLMYDPIF 300
301 ELNEKLSRSF LDTFFTRLQF KYPILDEQEI YTFYDHYLHN KILIPPSSPA TSSAAPPSNS 360
361 HSYSEIEFHF LSGRMWLVFS ISAYLLMTTG KYKGFPPHRY FSTAIRHITK CGLHLNYVQQ 420
421 IELLTLLVLY IIRTDRDSLI LYDIIKDVMG ISKKKLHLNQ WYPNDPFANK KLRLFWCVYL 480
481 LERMICVAVG KPYTIKESEI NLPLFNNDSF YTKGVHAAAP STNDHGVQFI NQSLKLRRIE 540
541 SQFVETLQLL KNDSRSVKQS IDQLPLVRKF FEDLEVWRKS YSTLDVKNFE NETLKLYYYR 600
601 SVRLLIQPYL EFFAPEDRLF RECQAAAGQI CQLYKIFHQK TLNGHSTPAV HTVFVAGVTL 660
661 IYCMWLARNF DDQRRKKLGD ASKHTRPLIS ASLFSTMDDL RACSVCLYVM TERSNFARTF 720
721 RDTFDQLMNA TVGNLIERCG PDSSELIFMA SSVAKRTEPK NINDEANKAI SSGDTLHDSN 780
781 SANAANLSNS NDKNISHNGG MPPAVARIFG KGQAEEHAGF VENSQVDLAE QEKFKKKQGV 840
841 LEKTSVPKSL AHLLTKMDDR SRISNSSMSY TTSSSSSSSS SSSSSTLSFP SSQEKNLKIN 900
901 VNNDNNGMTI SSVNREHNNN HNNNNDNNNN NNNNNNNSNN NNNVNNNDNE SNSRSTTNNS 960
961 CNNGNNSQYV RNNNVTMEND VERPIQDQYI VKKPTNQTEF DWQVFQQQAF LQQQLAQHNL1020
1021 QAYLSSLNTD TMTNRSPSKS SSISTASSHS DPIPIAMTQS PTPYPQTSNM LPQQHVSRPL1080
1081 PQQQREQPQQ HITSPQRFSE SNFTNQLNNG MINSNPLQSA IFSNHTSENK QLRDVEESNF1140
1141 STSPLRADYG NNIISSIPAS FTSNSIPVSV KQARNGSSSG DILFSNGAHD MINNISTWTN1200
1201 NSVLDALNSK SILQTIFPQS QEPSSLSMDK QQQQHQQQNM CSENNVTANN FQQTQNDPSY1260
1261 NRNLFMMSNQ EGVQYNLDET EKNGPKTQVE ANTSANLHFD NVIPTVTNAD IRKKRSNWDN1320
1321 MMTSGPVEDF WTINDDYGFL T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
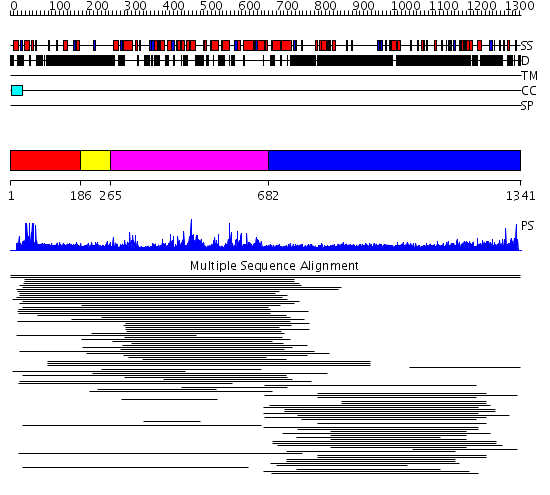
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | 14.522879 | PPR1 | |
| 2 | View Details | [186..264] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [265..681] | 4.08199 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [682..1341] | 15.144996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [505..732] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [733..1052] | 3.26 | 30S subunit | |
| 7 | View Details | [1053..1249] | 16.0 | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | |
| 8 | View Details | [1250..1341] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)