













| General Information: |
|
| Name(s) found: |
YEL1_YEAS8
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae EC1118 |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCASLNEVKK NDTYGVSQKG YNDNFSESEG VLHGSKSMPT SMKNMLQSPT MVNMCDILQN 60
61 KEAANDEKPV IPTTDTATAG TGTEDISSTQ SEETDQNSHL IASEILEGTF KDVSYKEYAN 120
121 FLGNDNNNQV LTEFVKLLSP LPSSLLETLF NLSKSIYFIA EAQNIDRILE CLSIEWIACH 180
181 PNTHWKSGYK SCHIVLFSLL ILNSDLHNNF QVDHKKIKFS MVAFINNTLR ALREENEYEE 240
241 LKIYSREHLI IEELSEYYKT LNETPLPLCT ESRTSINISD NQSSLKRFST LGSREFSTSN 300
301 LRSVNSNSTT LYSRDGQVSV REMSAKSNKN FHNNHPMDAL YLKESFDDGL ITENGSSWFM 360
361 DDLILISKKS LPRKYSKRDK DQVAAPKMTS KRNKSFFGWL KPSKTTTLIE HTSRRTSLSY 420
421 LNKDSEWERV KIQVKEGRIF IFKIKPDVKD IIQSSETDSA TIDYFKDISS SYFAYSLLEA 480
481 EAHVVQDNII IGSGAMKSNV CNKNTKRKSG NFTVSFPENI NGPKLVLEFQ TRSVEEAHKF 540
541 MDCINFWAGR ISPVPLTQFE AVSNAEYGWS DKILTEHASL NLKNIVVSEW KPLLGLELLY 600
601 EDAKDVEMVE LKERLKELMN FTRQLGIWID KHNEIKDKLV EIWSFDDNYF EAVMNNWNSR 660
661 YLYMNNQYKK RLSYLKALQK AMGSVQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
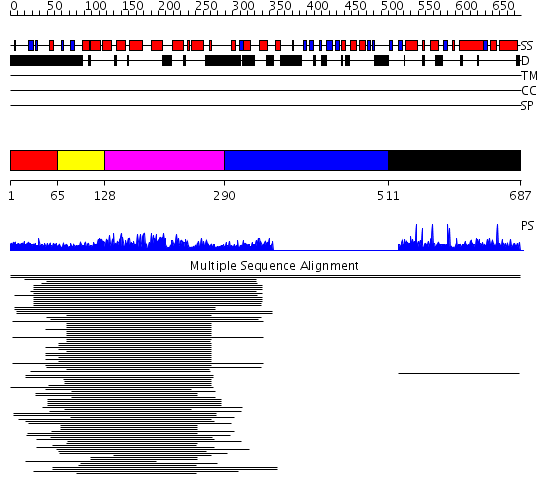
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [65..127] | 108.9897 | Cytohesin-1/b2-1 | |
| 3 | View Details | [128..289] | 108.9897 | Cytohesin-1/b2-1 | |
| 4 | View Details | [290..510] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [511..687] | 1.002941 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)