













| General Information: |
|
| Name(s) found: |
XRN2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1006 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVPSFFRWL SRKYPKIISP VLEEQPQIVD GVILPLDYSA SNPNGELDNL YLDMNGIVHP 60
61 CSHPENKPPP ETEDEMLLAV FEYTNRVLNM ARPRKVLVMA VDGVAPRAKM NQQRARRFRS 120
121 ARDAQIENEA REEIMRQREE VGEIIDDAVR NKKTWDSNAI TPGTPFMDKL AAALRYWTAF 180
181 KLATDPGWKN LQVIISDATV PGEGEHKIMN FIRSQRADPE YNPNTTHCIY GLDADLIFLG 240
241 LATHEPHFKI LREDVFAQDN RKRNNLKDTI NMTEEEKQFL QKQNSEQPFL WLHINVLREY 300
301 LSAELWVPGL PFTFDLERAI DDWVFMCFFC GNDFLPHLPC LDVRENSIDI LLDIWKVVLP 360
361 KLKTYMTCDG VLNLPSVETL LQHLGSREGD IFKTRHIQEA RKKEAFERRK AQKNMSKGQD 420
421 RHPTVATEQL QMYDTQGNLA KGSWNLTTSD MVRLKKELML ANEGNEEAIA KVKQQSDKNN 480
481 ELMKDISKEE IDDAVSKANK TNFNLAEVMK QKIINKKHRL EKDNEEEEIA KDSKKVKTEK 540
541 AESECDLDAE IKDEIVADVN DRENSETTEV SRDSPVHSTV NVSEGPKNGV FDTDEFVKLF 600
601 EPGYHERYYT AKFHVTPQDI EQLRKDMVKC YIEGVAWVLM YYYQGCASWN WFYPYHYAPL 660
661 ATDFHGFSHL EIKFEEGTPF LPYEQLMSVL PAASGHALPK IFRSLMSEPD SEIIDFYPEE 720
721 FPIDMNGKKM SWQGIALLPF IDQDRLLTAV RAQYPLLSDA ERARNIRGEP VLLISNKNAN 780
781 YERFSKKLYS KENNNNNVVV KFQHFKSGLS GIVSKDVEGF ELNGKIVCPI QGGSLPNLST 840
841 TLILKMSYRL IPLPSRNKSI ILNGFIPSEP VLTAYDLDSI MYKYNNQNYS RRWNFGNDLK 900
901 QNIVPVGPKG ITQYKPRTGG YRAFFYFAEL SRNNVQPAHN YGRNSYNSQP GFNNSRYDGG 960
961 NNNYRQNSNY RNNNYSGNRN SGQYSGNSYS RNNKQSRYDN SRANRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
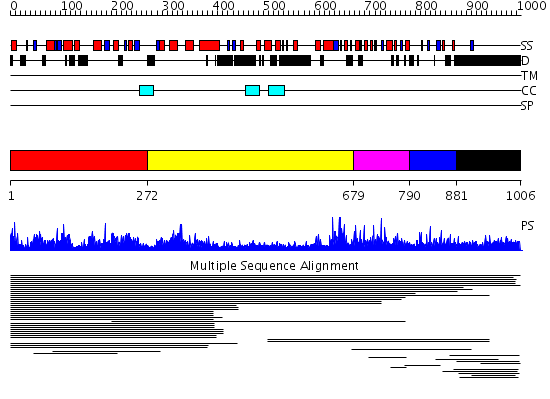
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..271] | 188.481486 | XRN 5'-3' exonuclease N-terminus No confident structure predictions are available. | |
| 2 | View Details | [272..678] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [679..789] | 1.015973 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [790..880] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [881..1006] | 2.012983 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)