













| General Information: |
|
| Name(s) found: |
XPOT_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1100 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLERIQQLVN AVNDPRSDVA TKRQAIELLN GIKSSENALE IFISLVINEN SNDLLKFYGL 60
61 STLIELMTEG VNANPNGLNL VKFEITKWLK FQVLGNKQTK LPDFLMNKIS EVLTTLFMLM 120
121 YSDCNGNQWN SFFDDLMSLF QVDSAISNTS PSTDGNILLG LEFFNKLCLM INSEIADQSF 180
181 IRSKESQLKN NNIKDWMRDN DIMKLSNVWF QCLKLDEQIV SQCPGLINST LDCIGSFISW 240
241 IDINLIIDAN NYYLQLIYKF LNLKETKISC YNCILAIISK KMKPMDKLAF LNMINLTNEL 300
301 TYYHQAISMN PQIITFDNLE VWESLTKLIT SFGIEFTIII EQVNDDQKLD TLYKQSVISN 360
361 VDSILLEKII PILLEFMNNE FDSITAKTFP FWSNYLAFLK KYKASSPNFV PLHKDFLDNF 420
421 QQICFKRMKF SDDEVTQDDF EEFNETVRFK LKNFQEIIVV IDPSLFLNNI SQEISANLMN 480
481 CKNESWQIFE LTIYQIFNLS ECTKNNYFGL NKNEIMTSQP SLTLVRFLNE LLMMKDFLLA 540
541 IDNEQIQILF MELIVKNYNF IFSTSANTAN ATDDDEKYLL ILNIFMSSFA MFNKRENVRL 600
601 RSWYLFTRFL KLTRINLKKI LFANKNLVNE ITNKISPLLH IKVTSINAQG TDDNDTIFDN 660
661 QLYIFEGIGF IITLNNSSQE LTAATANTPI DYDILDQILT PLFTQLEGCI TQGASPVVIL 720
721 ECHHILMAIG TLARGLHIGL VPENQVNNMV VNKKLINDSL IHKFSNIAEV ILVTFSFFNK 780
781 FENIRDASRF TFARLIPILS NKILPFINKL IELILSSTDL KSWEMIDFLG FLSQLIHMFH 840
841 TDTDCYQLFN QLLTPLINKV HSIIEEIDEQ HDQQSSSNKP IDTAVTATSV NKNIVVTDSY 900
901 RDKILLKKAY CTFLQSFTNN SVTSILLSDI NRAILPVILN DLVTYTPQEI QETSMMKVSL 960
961 NVLCNFIKCF GNGTCLDNDD INKDPNLKID GLNEYFIMKC VPIIFEIPFN PIYKFNIKEG1020
1021 NFKTMAYDLA RLLRELFIVS SNPTTNENEC VKYLTQIYLP QIQLPQELTI QLVNMLTTMG1080
1081 QKQFEKWFVD NFISVLKQGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
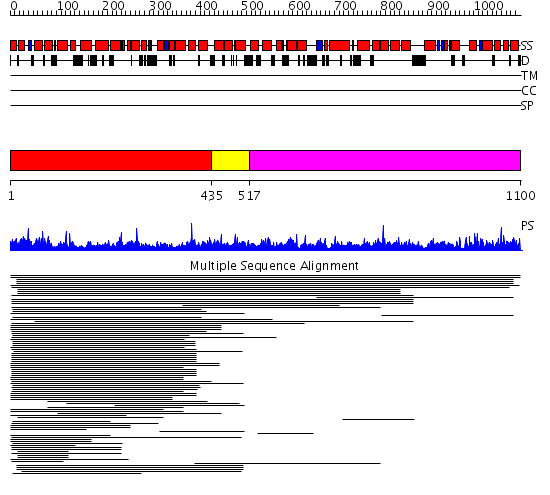
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..434] | 11.053947 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [435..516] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [517..1100] | 1.014975 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1013..1100] | 1.031986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)