













| General Information: |
|
| Name(s) found: |
WHI3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 661 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSSVYFDQT GSFASSSDNV VSSTTNTHNI SPSHRSSLNL NTTSHPHEAS GRGSASGELY 60
61 LNDTNSPLAI SSMLNTLALG SMPQDIASSN ISNHDNNIKG SYSLKLSNVA KDITLRECYA 120
121 IFALAEGVKS IELQKKNSSS SITSASLEDE NDIFIIARFE LLNLAINYAV ILNSKNELFG 180
181 PSFPNKTTVE IIDDTTKNLV SFPSSAIFND TSRLNKSNSG MKRPSLLSQR SRFSFSDPFS 240
241 NDSPLSQQQS QQQQQQPQQP QQHSTQKHSP QQCNQQQVNS SIPLSSQGQV IGLHSNHSHQ 300
301 DLSVESTIQT SDIGKSFLLR DNTEINEKIW GTSGIPSSIN GYMSTPQPST PTLEWGNTSA 360
361 SQHGSSFFLP SAASTAIAPT NSNTSANANA SSNNGASNNG ANQALSASSQ QPMMQIGNTI 420
421 NTSLTSSNSL PPYGLMSSQS QHISNMVNTS DMNITPQKQN RFMQQPQPEH MYPVNQSNTP 480
481 QKVPPARLSS SRNSHKNNST TSLSSNITGS ASISQADLSL LARIPPPANP ADQNPPCNTL 540
541 YVGNLPSDAT EQELRQLFSG QEGFRRLSFR NKNTTSNGHS HGPMCFVEFD DVSFATRALA 600
601 ELYGRQLPRS TVSSKGGIRL SFSKNPLGVR GPNSRRGGSG NPNPNVNMLS SYNSNVGHIK 660
661 N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
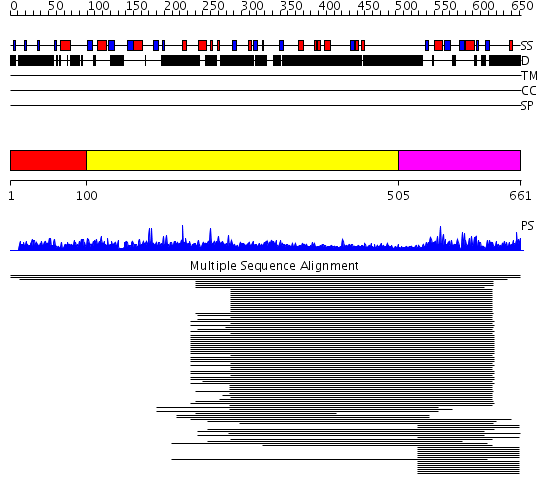
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..504] | 10.98 | RBP1 | |
| 3 | View Details | [505..661] | 39.522879 | Poly(A)-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)