













| General Information: |
|
| Name(s) found: |
VIP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1146 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGIKKEPIE SDEVPQQETK NNLPSAPSEM SPLFLNKNTQ KAMQSIAPIL EGFSPKTSAS 60
61 ENMSLKLPPP GIQDDHSEEN LTVHDTLQRT ISTALGNGNN TNTVTTSGLK KADSESKSEA 120
121 DPEGLSNSNI VNDADNINSI SKTGSPHLPQ GTMDAEQTNM GTNSVPTSSA SSRKSSTSHP 180
181 KPRLPKVGKI GVCAMDAKVL SKPMRHILNR LIEHGEFETV IFGDKVILDE RIENWPTCDF 240
241 LISFFSSGFP LDKAIKYVKL RKPFIINDLI MQKILWDRRL CLQVLEAYNV PTPPRLEISR 300
301 DGGPRANEEL RAKLREHGVE VKPVEEPEWK MVDDDTLEVD GKTMTKPFVE KPVDGEDHNI 360
361 YIYYHSKNGG GGRRLFRKVG NKSSEFDPTL VHPRTEGSYI YEQFMDTDNF EDVKAYTIGE 420
421 NFCHAETRKS PVVDGIVRRN THGKEVRYIT ELSDEEKTIA GKVSKAFSQM ICGFDLLRVS 480
481 GKSYVIDVNG FSFVKDNKAY YDSCANILRS TFIEAKKKMD MEKKNLPIIR EEKEQKWVFK 540
541 GLAIIIRHAD RTPKQKFKHS FTSPIFISLL KGHKEEVVIR NVNDLKIVLQ ALRIALDEKA 600
601 GNPAKIKVLA NALEKKLNFP GTKIQLKPVL NKENEVEKVQ FILKWGGEPT HSAKYQATEL 660
661 GEQMRQDFDL LNKSILQNIK IFSSSERRVL HTAQYWTRAL FGADELGSDE ISIRKDLLDD 720
721 SNAAKDLMDK VKKKLKPLLR EGKEAPPQFA WPSKMPEPYL VIKRVVELMN YHKKIMDNNF 780
781 AKKDVNSMQT RWCTSEDPSL FKERWDKLFK EFNNAEKVDP SKISELYDTM KYDALHNRQF 840
841 LENIFDPGLP NEAIADELGS HSLVDRYPIN VLAKNNFKII DSHSMNNSGK NSSNSVGSLG 900
901 WVLESGKTST ARNPKSSSQF DEPRFMQLRE LYKLAKVLFD FICPKEYGIS DAEKLDIGLL 960
961 TSLPLAKQIL NDIGDMKNRE TPACVAYFTK ESHIYTLLNI IYESGIPMRI ARNALPELDY1020
1021 LSQITFELYE STDASGQKSH SIRLKMSPGC HTQDPLDVQL DDRHYISCIP KISLTKHLDM1080
1081 DYVQQKLRNK FTRVIMPPKF TPVNITSPNL SFQKRKTRRK SVSVEKLKRP ASSGSSSSTS1140
1141 VNKTLD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
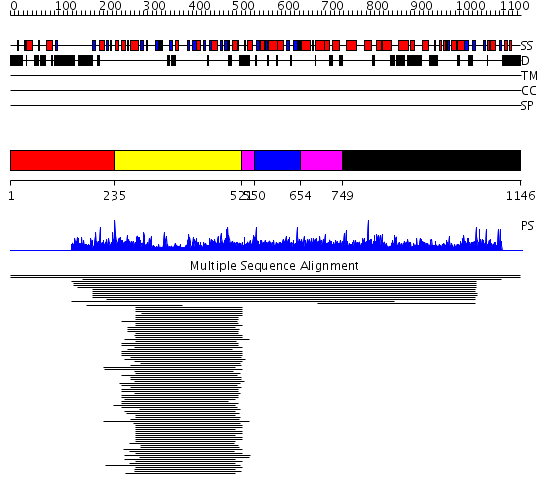
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [235..520] | 41.522879 | D-Ala-D-Ala ligase, N-domain; D-ala-D-ala ligase, C-domain | |
| 3 | View Details | [521..549] [654..748] |
8.82 | Phytase (myo-inositol-hexakisphosphate-3-phosphohydrolase) | |
| 4 | View Details | [550..653] | 8.82 | Phytase (myo-inositol-hexakisphosphate-3-phosphohydrolase) | |
| 5 | View Details | [749..1146] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)