













| General Information: |
|
| Name(s) found: |
VAC14_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 880 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKSIAKGLS DKLYEKRKAA ALELEKLVKQ CVLEGDYDRI DKIIDELCRD YAYALHQPMA 60
61 RNAGLMGLAA TAIALGINDV GRYLRNILPP VLACFGDQND QVRFYACESL YNIAKIAKGE 120
121 ILVYFNEIFD VLCKISADTE NSVRGAAELL DRLIKDIVAE RASNYISIVN NGSHGLLPAI 180
181 KTDPISGDVY QEEYEQDNQL AFSLPKFIPL LTERIYAINP DTRVFLVDWL KVLLNTPGLE 240
241 LISYLPSFLG GLFTFLGDSH KDVRTVTHTL MDSLLHEVDR ISKLQTEIKM KRLERLKMLE 300
301 DKYNNSSTPT KKADGALIAE KKKTLMTALG GLSKPLSMET DDTKLSNTNE TDDERHLTSQ 360
361 EQLLDSEATS QEPLRDGEEY IPGQDINLNF PEVITVLVNN LASSEAEIQL IALHWIQVIL 420
421 SISPNVFIPF LSKILSVLLK LLSDSDPHIT EIAQLVNGQL LSLCSSYVGK ETDGKIAYGP 480
481 IVNSLTLQFF DSRIDAKIAC LDWLILIYHK APNQILKHND SMFLTLLKSL SNRDSVLIEK 540
541 ALSLLQSLCS DSNDNYLRQF LQDLLTLFKR DTKLVKTRAN FIMRQISSRL SPERVYKVIS 600
601 SILDNYNDTT FVKMMIQILS TNLITSPEMS SLRNKLRTCE DGMFFNSLFK SWCPNPVSVI 660
661 SLCFVAENYE LAYTVLQTYA NYELKLNDLV QLDILIQLFE SPVFTRMRLQ LLEQQKHPFL 720
721 HKCLFGILMI IPQSKAFETL NRRLNSLNIW TSQSYVMNNY IRQRENSNFC DSNSDISQRS 780
781 VSQSKLHFQE LINHFKAVSE EDEYSSDMIR LDHGANNKSL LLGSFLDGID EDKQEIVTPI 840
841 SPMNEAINEE MESPNDNSSV ILKDSGSLPF NRNVSDKLKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
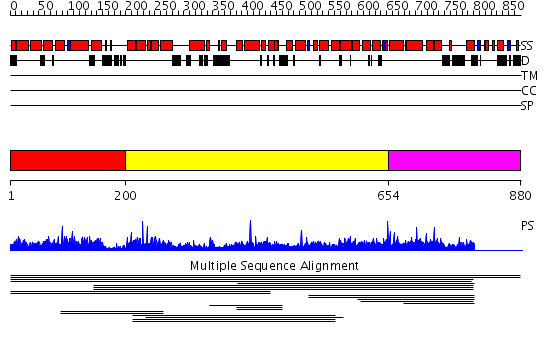
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [200..653] | 11.67 | Clathrin heavy chain proximal leg segment | |
| 3 | View Details | [654..880] | 1.009959 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [787..880] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)