













| General Information: |
|
| Name(s) found: |
UBP3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 912 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMQDANKEE SYSMYPKTSS PPPPTPTNMQ IPIYQAPLQM YGYTQAPYLY PTQIPAYSFN 60
61 MVNQNQPIYH QSGSPHHLPP QNNINGGSTT NNNNINKKKW HSNGITNNNG SSGNQGANSS 120
121 GSGMSYNKSH TYHHNYSNNH IPMMASPNSG SNAGMKKQTN SSNGNGSSAT SPSYSSYNSS 180
181 SQYDLYKFDV TKLKNLKENS SNLIQLPLFI NTTEAEFAAA SVQRYELNMK ALNLNSESLE 240
241 NSSVEKSSAH HHTKSHSIPK HNEEVKTETH GEEEDAHDKK PHASKDAHEL KKKTEVKKED 300
301 AKQDRNEKVI QEPQATVLPV VDKKEPEESV EENTSKTSSP SPSPPAAKSW SAIASDAIKS 360
361 RQASNKTVSG SMVTKTPISG TTAGVSSTNM AAATIGKSSS PLLSKQPQKK DKKYVPPSTK 420
421 GIEPLGSIAL RMCFDPDFIS YVLRNKDVEN KIPVHSIIPR GIINRANICF MSSVLQVLLY 480
481 CKPFIDVINV LSTRNTNSRV GTSSCKLLDA CLTMYKQFDK ETYEKKFLEN ADDAEKTTES 540
541 DAKKSSKSKS FQHCATADAV KPDEFYKTLS TIPKFKDLQW GHQEDAEEFL THLLDQLHEE 600
601 LISAIDGLTD NEIQNMLQSI NDEQLKVFFI RNLSRYGKAE FIKNASPRLK ELIEKYGVIN 660
661 DDSTEENGWH EVSGSSKRGK KTKTAAKRTV EIVPSPISKL FGGQFRSVLD IPNNKESQSI 720
721 TLDPFQTIQL DISDAGVNDL ETAFKKFSEY ELLPFKSSSG NDVEAKKQTF IDKLPQVLLI 780
781 QFKRFSFINN VNKDNAMTNY NAYNGRIEKI RKKIKYGHEL IIPEESMSSI TLKNNTSGID 840
841 DRRYKLTGVI YHHGVSSDGG HYTADVYHSE HNKWYRIDDV NITELEDDDV LKGGEEASDS 900
901 RTAYILMYQK RN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
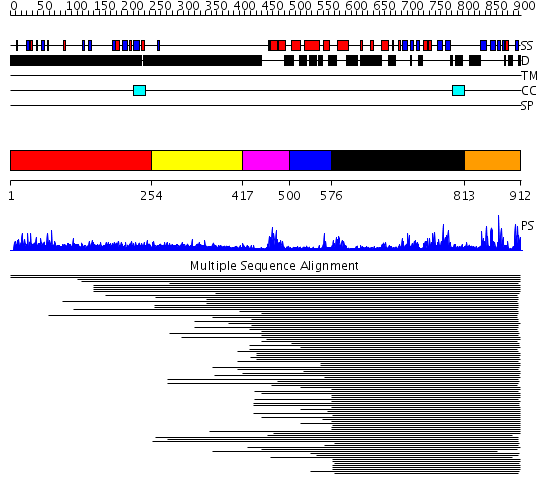
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..253] | 14.84 | RBP1 | |
| 2 | View Details | [254..416] | 15.0 | No description for 1ldvA was found. | |
| 3 | View Details | [417..499] | 9.79588 | No description for PF00442 was found. No confident structure predictions are available. | |
| 4 | View Details | [500..575] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [576..812] | 4.22699 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [813..912] | 24.267606 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)