













| General Information: |
|
| Name(s) found: |
UBP11_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLNPDQILN LVRKVYEVDI KQFYSQLRLK NLRGLLDHAA HLFNVYLRDL EINQEMEALT 60
61 AFIIGCYYLY LIIPQSLQFQ TRNNLYSSYA KLKNDYQDEH VMGYVLKVVR DESTVIVDRY 120
121 LAESNGICRT IKRKRAYSLP LRPLPVHMAS LSIHNKFDGS LHEIPNELTK PTNDNSKEDI 180
181 VRESNQIASS NKLEAGSEVA YYTSKEALSK PSYLKLSTGK DALFKTLSSP ATAPPVHSLE 240
241 VSSQIRDSSQ DSSSSLSKVE KPKEEEGKIE AIESSAPKAY NLPVIEDSND LLSELSITGL 300
301 QNPCNTCYIN SIIQCLFGTT LFRDLFLTKK YRLFLNTNKY PKEVQLSRSI YVLFKKMYLN 360
361 GGRAIIPNRF LKMCKKLRPD LNIPDDQQDT QEFLLIVLAR IHEELSNENV VKYYPDLVSY 420
421 DANALQVNPS KYEKWYERNV ITDGLSPIDH IYRGQLENIL KCQRCGNSSY SYSTFYVLSL 480
481 AIPKLSLYSF TSKSRKIKLE DCINLFTGDE ELSGDNAWDC PNCRITDSKS KKEEITSQKK 540
541 KSTIFGFHSR SRSKSPHHHH HHHHSSDDST KNAKKRNSKK LTTIKSLDFI VLPPILVIHL 600
601 SRFYYDLTKK NSTVITYPLI LNIILKNGKV IRYKLYGTVN HSGNLINGHY TSVVNKEKSH 660
661 EIGLNRQVWV TFDDDYIQQH RKDRNNFEAG KTEMSSDEVY VLFYERMDEE NYEEEFC |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
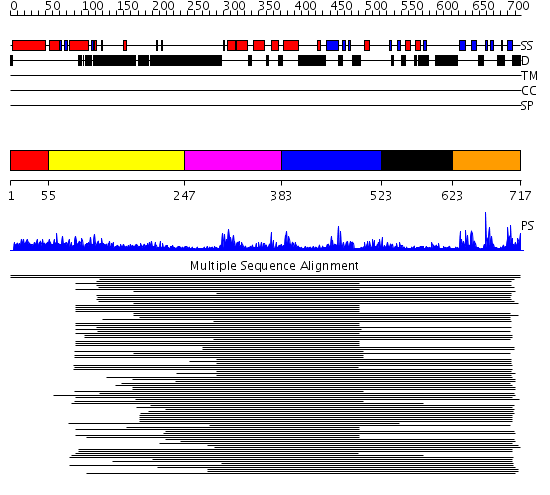
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..54] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [55..246] | 1.14798 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [247..382] | 13.327902 | No description for PF00442 was found. No confident structure predictions are available. | |
| 4 | View Details | [383..522] | 2.236988 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [523..622] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [623..717] | 23.070581 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)