













| General Information: |
|
| Name(s) found: |
THDH_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 576 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSATLLKQPL CTVVRQGKQS KVSGLNLLRL KAHLHRQHLS PSLIKLHSEL KLDELQTDNT 60
61 PDYVRLVLRS SVYDVINESP ISQGVGLSSR LNTNVILKRE DLLPVFSFKL RGAYNMIAKL 120
121 DDSQRNQGVI ACSAGNHAQG VAFAAKHLKI PATIVMPVCT PSIKYQNVSR LGSQVVLYGN 180
181 DFDEAKAECA KLAEERGLTN IPPFDHPYVI AGQGTVAMEI LRQVRTANKI GAVFVPVGGG 240
241 GLIAGIGAYL KRVAPHIKII GVETYDAATL HNSLQRNQRT PLPVVGTFAD GTSVRMIGEE 300
301 TFRVAQQVVD EVVLVNTDEI CAAVKDIFED TRSIVEPSGA LSVAGMKKYI STVHPEIDHT 360
361 KNTYVPILSG ANMNFDRLRF VSERAVLGEG KEVFMLVTLP DVPGAFKKMQ KIIHPRSVTE 420
421 FSYRYNEHRH ESSSEVPKAY IYTSFSVVDR EKEIKQVMQQ LNALGFEAVD ISDNELAKSH 480
481 GRYLVGGASK VPNERIISFE FPERPGALTR FLGGLSDSWN LTLFHYRNHG ADIGKVLAGI 540
541 SVPPRENLTF QKFLEDLGYT YHDETDNTVY QKFLKY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
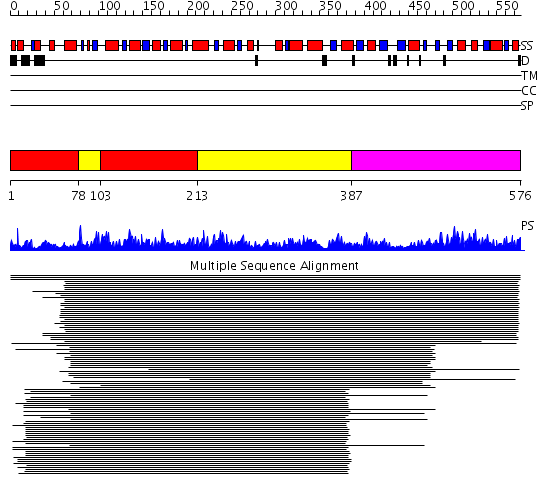
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] [103..212] |
1441.0 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain | |
| 2 | View Details | [78..102] [213..386] |
1441.0 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain | |
| 3 | View Details | [387..576] | 1441.0 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)