













| General Information: |
|
| Name(s) found: |
TAF2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1407 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMSFSKNATP RAIVSESSTL HEMKFRNFRV AHEKISLDID LATHCITGSA TIIIIPLIQN 60
61 LEYVTFDCKE MTIKDVLVEN RRCDQFIHDD PLQTNLNGLT SQNVLYSDNS IEQSHFLRSK 120
121 FASLNEYPET DSKSQLTIKI PSSIKISLED ANALSNYTPI TPSIKTTPGF QESVFTPITL 180
181 QIEYEIRNPK SGIKFDTVYA DKPWLWNVYT SNGEICSSAS YWVPCVDLLD EKSTWELEFS 240
241 VPRLVKNIGT SKLIGQNGEE SEKEKEDTPE HDEEEEGKPA RVIKDEDKDS NLKNDEEGKN 300
301 SKSKDAQDND EEEEEGESDE EEEEGEEERR NIEESNNPSL RDVIVCCSEY SNIKELPHPI 360
361 DLTKKKCIFQ IINPVAPHHI GWAIGAFNSW SLPLISPPSV DAEDEVEEDK LRENVVDNVN 420
421 DTMDDDIGSD IIPIQIFTLP TQETDELTVI NSTVVCQKII DFYSKEFGSY PFTCYSMVFL 480
481 PTAPSKHMDF AALGICNTRL LYPLEVIDKA FSTTNELAWA LANQWSCVNI TPLDMNDYWC 540
541 CLGIAGYMVF QVTKKLMGNN TYKYQLKRNS EAIVEQDFEK PPIGSTFTGS SRPISWSSKD 600
601 LSFIQLKAPM ILHILDRRMT KTERSFGMSR VLPKIFLQAM SGDLPNNSLT SSHFQHVCER 660
661 VNKSKLENFF NEWVYGSGVP ILRVTQRFNR KRMVIELGIR QVQDEELGHE KVVGEEGFFK 720
721 SALDHLEHPD LNRTECFTGS MTIRIHEHDG TPYEHIVEIK DTFTKIDIQY NTKYRRLRKR 780
781 GGGANDENGV ENNNEEKPIV VDVNCLGNVY MSPEECSRFS LTEFNRTSES NELLKQNEAF 840
841 EWIRIDSDLE WICQMHINQP DYMFSSQLRQ DGDIEAQLEA IRYYEDVVVN GGVKSLVYSS 900
901 ILFRTAIDER YFFGIRLAAC EALSKYVYDP DFTGGVKHLI QIFQILFCLE DSNIPKSNNF 960
961 ENPKLYFLQC NIPKYLAKVK NENGKCPKLV KQFLLDILVY NENGENKYSD DAYVRSLIEN1020
1021 VVKVALNEYK DKAYMEKVKT QLLRYENLVN WLSSYESLIK TTIMYAKYKL HKVGAYDFTE1080
1081 LTGMIMHTLT LGINNGDISR ESFQNEFLMV LKIMLLEGGL KNKDALVLFT EILCFHEDSY1140
1141 IRDKSVDVLS ECVNLVVMDG SLDTISDDIK SSVQSVHNEV KNIKSEDDIE LFLSGHYVDD1200
1201 MKIKIEKIGR QNISGLIQIC RDMFKGYSPL KILLWDVLNL PVLSLYQRKQ IHDLVRVMYT1260
1261 LINSFVVRLE TPRERRLVAK MNSNEEGKLD IVIKRESILK VHIKKEVTST VEAPKKANKI1320
1321 KISLKGDKPV RKVEKQIVKP KVTSKQRKVK SHVNRMGSLP LRFVKIQQQP RVMVHLSSVP1380
1381 YSQFVQITKV TSRSFMVKIR TKNDAKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
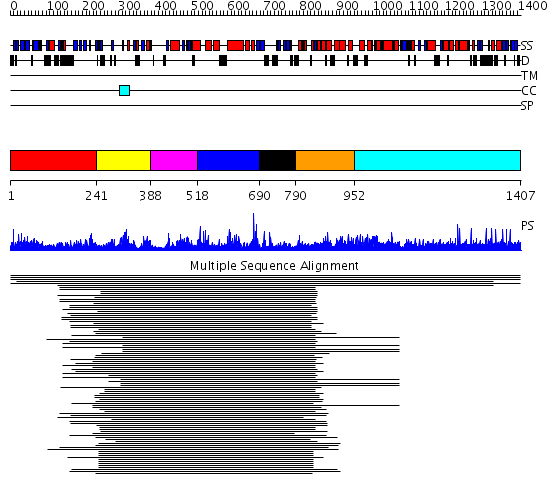
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | 11.15 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 2 | View Details | [241..387] | 80.0 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 3 | View Details | [388..517] | 80.0 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 4 | View Details | [518..689] | 80.0 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 5 | View Details | [690..789] | 80.0 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 6 | View Details | [790..951] | 2.127996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [952..1407] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)