













| General Information: |
|
| Name(s) found: |
TAF1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1066 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKQQGSGKT NLANEDEAYE AIFGGEFGSL EIGSYIGGDE GANSKDYTEH LPDAVDFEDE 60
61 DELADDDDDL PEESDANLHP AMMTMGAYDD VNENGAVLGI DSNSLNMQLP EINGDLSQQF 120
121 ILEDDGGTPA TSNALFMGMD ANEIHLATET GVLDGSGANE IGHSQLSIGG VNGNDMSING 180
181 GFIMEPDMSD GKHKKATKLD LINHEKYLLK KYFPDFEKGK ILKWNKLIYR RSVPYHWHSE 240
241 ISRVKKPFMP LNLKFKVQQD DKRLFNSRTI SYVAPIYQGK NNLLQSNSSA SRRGLIHVSI 300
301 DELFPIKEQQ KKRKIIHDEK TISEDLLIAT DDWDQEKIIN QGTSSTATLA DSSMTPNLKF 360
361 SGGYKLKSLI EDVAEDWQWD EDMIIDAKLK ESKHAELNMN DEKLLLMIEK TNNLAQQKQQ 420
421 LDSSNLILPL NETILQQKFN LSNDDKYQIL KKTHQTKVRS TISNLNIQHS QPAINLQSPF 480
481 YKVAVPRYQL RHFHRENFGS HIRPGTKIVF SKLKARKRKR DKGKDVKESF STSQDLTIGD 540
541 TAPVYLMEYS EQTPVALSKF GMANKLINYY RKANEQDTLR PKLPVGETHV LGVQDKSPFW 600
601 NFGFVEPGHI VPTLYNNMIR APVFKHDISG TDFLLTKSSG FGISNRFYLR NINHLFTVGQ 660
661 TFPVEEIPGP NSRKVTSMKA TRLKMIIYRI LNHNHSKAIS IDPIAKHFPD QDYGQNRQKV 720
721 KEFMKYQRDG PEKGLWRLKD DEKLLDNEAV KSLITPEQIS QVESMSQGLQ FQEDNEAYNF 780
781 DSKLKSLEEN LLPWNITKNF INSTQMRAMI QIHGVGDPTG CGEGFSFLKT SMKGGFVKSG 840
841 SPSSNNNSSN KKGTNTHSYN VAQQQKAYDE EIAKTWYTHT KSLSISNPFE EMTNPDEINQ 900
901 TNKHVKTDRD DKKILKIVRK KRDENGIIQR QTIFIRDPRV IQGYIKIKEQ DKEDVNKLLE 960
961 EDTSKINNLE ELEKQKKLLQ LELANLEKSQ QRRAARQNSK RNGGATRTEN SVDNGSDLAG1020
1021 VTDGKAARNK GKNTTRRCAT CGQIGHIRTN KSCPMYSSKD NPASPK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
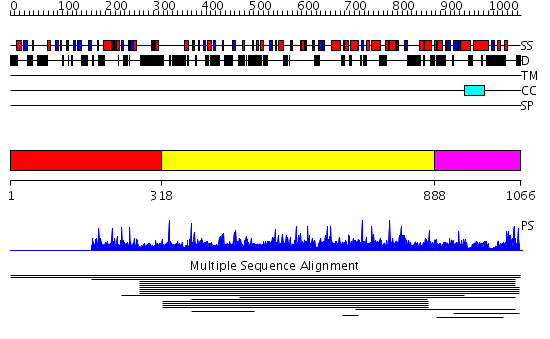
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [318..887] | 2.011837 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [888..1066] | 1.008972 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)