













| General Information: |
|
| Name(s) found: |
SRO7_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1033 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGSKRLKNV KEAFKSLKGQ NSETPIENSK ASFKSKNSKT STISKDAKSS SSLKIPISSN 60
61 NKNKIFSLAE TNKYGMSSKP IAAAFDFTQN LLAIATVTGE VHIYGQQQVE VVIKLEDRSA 120
121 IKEMRFVKGI YLVVINAKDT VYVLSLYSQK VLTTVFVPGK ITSIDTDASL DWMLIGLQNG 180
181 SMIVYDIDRD QLSSFKLDNL QKSSFFPAAR LSPIVSIQWN PRDIGTVLIS YEYVTLTYSL 240
241 VENEIKQSFI YELPPFAPGG DFSEKTNEKR TPKVIQSLYH PNSLHIITIH EDNSLVFWDA 300
301 NSGHMIMART VFETEINVPQ PDYIRDSSTN AAKISKVYWM CENNPEYTSL LISHKSISRG 360
361 DNQSLTMIDL GYTPRYSITS YEGMKNYYAN PKQMKIFPLP TNVPIVNILP IPRQSPYFAG 420
421 CHNPGLILLI LGNGEIETML YPSGIFTDKA SLFPQNLSWL RPLATTSMAA SVPNKLWLGA 480
481 LSAAQNKDYL LKGGVRTKRQ KLPAEYGTAF ITGHSNGSVR IYDASHGDIQ DNASFEVNLS 540
541 RTLNKAKELA VDKISFAAET LELAVSIETG DVVLFKYEVN QFYSVENRPE SGDLEMNFRR 600
601 FSLNNTNGVL VDVRDRAPTG VRQGFMPSTA VHANKGKTSA INNSNIGFVG IAYAAGSLML 660
661 IDRRGPAIIY MENIREISGA QSACVTCIEF VIMEYGDDGY SSILMVCGTD MGEVITYKIL 720
721 PASGGKFDVQ LMDITNVTSK GPIHKIDAFS KETKSSCLAT IPKMQNLSKG LCIPGIVLIT 780
781 GFDDIRLITL GKSKSTHKGF KYPLAATGLS YISTVEKNND RKNLTVIITL EINGHLRVFT 840
841 IPDFKEQMSE HIPFPIAAKY ITESSVLRNG DIAIRVSEFQ ASLFSTVKEQ DTLAPVSDTL 900
901 YINGIRIPYR PQVNSLQWAR GTVYCTPAQL NELLGGVNRP ASKYKESIIA EGSFSERSSD 960
961 DNNANHPEHQ YTKPTRKGRN SSYGVLRNVS RAVETRWDAV EDRFNDYATA MGETMNEAVE1020
1021 QTGKDVMKGA LGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
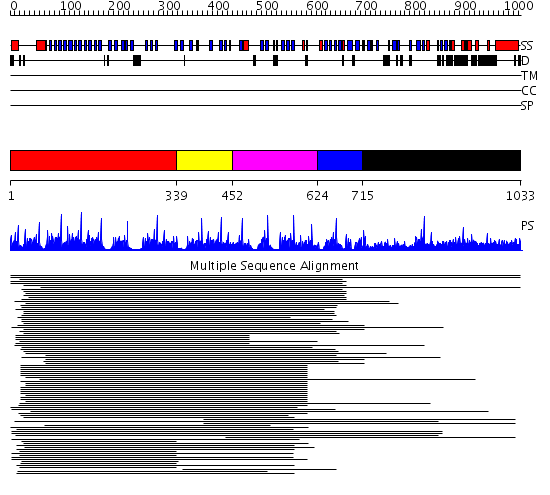
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..338] | 64.69897 | Tup1, C-terminal domain | |
| 2 | View Details | [339..451] | 2.522879 | TolB, C-terminal domain; TolB, N-terminal domain | |
| 3 | View Details | [452..623] | 9.198996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [624..714] | 3.379997 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [715..1033] | 1.15595 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)