













| General Information: |
|
| Name(s) found: |
SPT16_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1035 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEELNIDFDV FKKRIELLYS KYNEFEGSPN SLLFVLGSSN AENPYQKTTI LHNWLLSYEF 60
61 PATLIALVPG KVIIITSSAK AKHLQKAIDL FKDPESKITL ELWQRNNKEP ELNKKLFDDV 120
121 IALINSAGKT VGIPEKDSYQ GKFMTEWNPV WEAAVKENEF NVIDISLGLS KVWEVKDVNE 180
181 QAFLSVSSKG SDKFMDLLSN EMVRAVDEEL KITNAKLSDK IENKIDDVKF LKQLSPDLSA 240
241 LCPPNYKFNF DLLDWTYSPI IQSGKKFDLR VSARSTNDQL YGNGCILASC GIRYNNYCSN 300
301 ITRTFLIDPS EEMANNYDFL LTLQKEIVTN ILKPGRTPKE VYESVIEYIE KTKPELVPNF 360
361 TKNIGSLIGL EFRDSNFILN VKNDYRKIQR GDCFNISFGF NNLKDSQSAN NYALQLADTV 420
421 QIPLDETEPP RFLTNYTKAK SQISFYFNNE EEDNNKKKSS PATKVPSKPD RNSKILRTKL 480
481 RGEARGGAED AQKEQIRKEN QKKLHEKLEK NGLLRFSAAD ANGPDSEPRQ YFKKYESYVR 540
541 DSQLPTNIRD LRIHVDWKSQ TIILPIYGRP VPFHINSYKN GSKNEEGEYT YLRLNFNSPG 600
601 SSGGISKKVE ELPYEESADN QFVRSITLRS KDGDRMSETF KQIADLKKEA TKREQERKAL 660
661 ADVVQQDKLI ENKTGRTKRL DQIFVRPNPD TKRVPSTVFI HENGIRFQSP LRTDSRIDIL 720
721 FSNIKNLIFQ SCKGELIVVI HIHLKNPILM GKKKIQDVQF YREASDMSVD ETGGGRRGQS 780
781 RFRRYGDEDE LEQEQEERRK RAALDKEFKY FADAIAEASN GLLTVENTFR DLGFQGVPNR 840
841 SAVFCMPTTD CLVQLIEPPF LVINLEEVEI CILERVQFGL KNFDMVFVYK DFNKPVTHIN 900
901 TVPIESLDFL KQWLTDMDIP YTVSTINLNW ATIMKSLQDD PYQFFLDGGW NFLATGSDDE 960
961 ASDESEEEVS EYEASEDDVS DESAFSEDEE GSEVDDDISG DESEDYTGDE SEEGEDWDEL1020
1021 EKKAARADRG ANFRD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
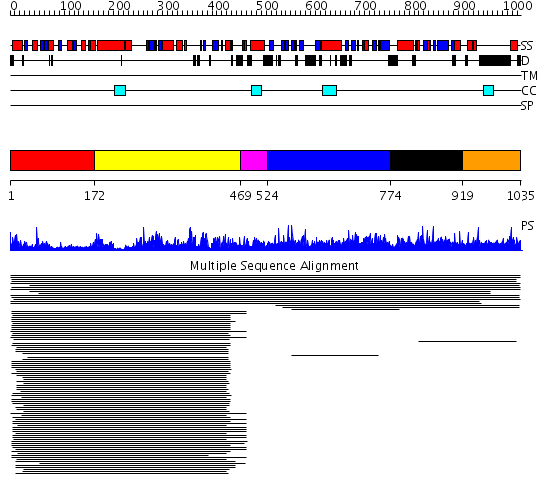
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 80.045757 | Aminopeptidase P; Aminopeptidase P, C-terminal domain | |
| 2 | View Details | [172..468] | 80.045757 | Aminopeptidase P; Aminopeptidase P, C-terminal domain | |
| 3 | View Details | [469..523] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [524..773] | 1.016941 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [774..918] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [919..1035] | 3.018996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)