













| General Information: |
|
| Name(s) found: |
SPH1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 661 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISSELTNDQ IIDLISDYKN FKRIIEASVP EDDRRRNQNR QNRNKDLTKL SNVQFWQLTT 60
61 DVNDELMKRL TDSGADASLN DLDLKRGKAQ SKLSRLKDAK FHKLILDIFT EIERRNLHHL 120
121 DMGTHNNGLD EGDLNFYLND TLFESFKIND DFMSVNGIIS IEVFRELKTQ FTLYFQNTLH 180
181 RIDPVDTTTT RLPILLETII KIAKLIGDLL PVLSSVSLQS SLENEIVYLK SALSHAITST 240
241 RYFLTYGDLI PRIVAQSSIS EVIFAFCNIA QIVKIKSTSR DDISRNEGEL SDIEAGMKPL 300
301 KIIEKVKNEK NGKDISSLGD SGSTAVSFPS SGKPITKKSD MPVVVASPSI SIIEKSESSI 360
361 RESGKVRNNT SGETNLASVS PLKNTKNSSR ITSEPSPREG LPLKVVSNSR SPSPQGNTLP 420
421 LIGKFRQDYQ ASPPKKVITK PVAETAKPYA NIPPAADVLY SPTVTKMRKF REKVQKFAPN 480
481 SGLGLRISTS EENLNNSDVN STTHNANINN LVEFVESKSM VVLPMAQGIL NDVQASKSKL 540
541 FKSARSVSRL CQNSIEIIPI LESVIDMTTK AMVQKSFKLD LGEHCKEIIE RLTDCSQKLS 600
601 ELCTYGCDST KLGKKRFYQK LADILMEVTK RTRELVECVK MANRQTLSQD LSSFFNYRSV 660
661 Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
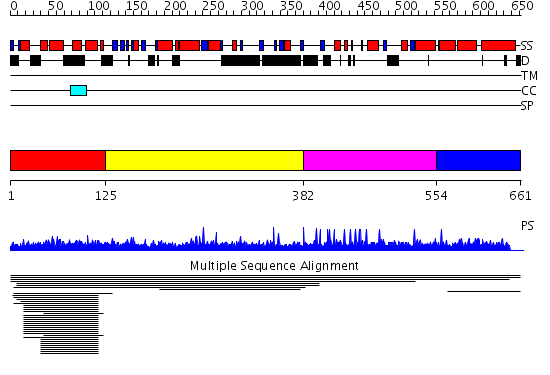
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 9.01398 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [125..381] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [382..553] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [554..661] | 1.002953 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)