













| General Information: |
|
| Name(s) found: |
SEN1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 2231 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSNNPDNNN SNNINNNNKD KDIAPNSDVQ LATVYTKAKS YIPQIEQVYQ GTNPNIQEAK 60
61 LLGELLQVLA EVPKGTHLFC DPILEPISIF SLTIFSFNEE ATATWLKNHF NPILSVCDKC 120
121 ILNFARGKCK MLQHFAIQRH VPHEHVAKFN DIVCQWRVEA VFPILRNISV NDNTGINITN 180
181 EIETAMYECL CNPHMLRLNK QLKATFEAIF KFFYDTKHRL LDVTNPLSIK TFISGVIFCW 240
241 CEGSKEENEW SRAFLKDLYS RNFHINLSNL TPDIIEEVYI HILFLQNPAN WTEIVVSQFW 300
301 SRLLPVFNLF DKDVFIEYFQ VPKNVESLKK TFKFPLEPIF KMWYNHLSKS YHDKPLDFLL 360
361 RGLTMFLNKF GSEFWSKIEP FTFHSILDII FNRDSFPIKL IKIQDNPIVE HQTEVYFQLT 420
421 GSVTDLLSWT LPFYHALSPS KRIQMVRKVS MAFLRIIANY PSLKSIPKAC LMNSATALLR 480
481 AVLTIKENER AMLYKNDEFE TVLLTKTDSR ALLNNPLIQD IIIRSASNPN DFYPGLGAAS 540
541 ASVATSTMMV LAECIDFDIL LLCHRTFKLY SGKPISEIPI STNVLENVTN KIDLRSFHDG 600
601 PLLAKQLLVS LKNINGLLIV PSNTAVAEAH NALNQKFLLL STRLMEKFAD ILPGQLSKIL 660
661 ADEDASQGFW SCIFSSDKHL YQAATNILYN TFDVEGRLEG ILAILNSNLT VNLKNINVML 720
721 QRLINCEFYE PCPRAVRVLM DVVSAFVDPI SGVFANFQTL KSQNTEKEFL KFWESCWLFL 780
781 DTIYKFTLKW ASKYDYSELE NFTKDTLDLS RSLVDSFREF SDILHDQTKN LLLNVLETFK 840
841 NMLYWLRLSD EVLLESCVRL IISTSDLAHE KHVKVDDSLV EMMAKYASKA KRFSNKLTEQ 900
901 QASEILQKAK IFNKALTEEV ATEAENYRKE KELSRLGKVI DLTDSVPASP SLSPSLSSTI 960
961 ASSSAESRAD YLQRKALSSS ITGRPRVAQP KITSFGTFQS SANAKLHRTK PVKPLSKMEL1020
1021 ARMQLLNNRV VHPPSAPAFH TKSRGLSNKN DDSSSEESDN DIESARELFA IAKAKGKGIQ1080
1081 TVDINGKVVK RQTAAELAKQ ELEHMRKRLN VDMNPLYEII LQWDYTRNSE YPDDEPIGNY1140
1141 SDVKDFFNSP ADYQKVMKPL LLLESWQGLC SSRDREDYKP FSIIVGNRTA VSDFYDVYAS1200
1201 VAKQVIQDCG ISESDLIVMA YLPDFRPDKR LSSDDFKKAQ HTCLAKVRTL KNTKGGNVDV1260
1261 TLRIHRNHSF SKFLTLRSEI YCVKVMQMTT IEREYSTLEG LEYYDLVGQI LQAKPSPPVN1320
1321 VDAAEIETVK KSYKLNTSQA EAIVNSVSKE GFSLIQGPPG TGKTKTILGI IGYFLSTKNA1380
1381 SSSNVIKVPL EKNSSNTEQL LKKQKILICA PSNAAVDEIC LRLKSGVYDK QGHQFKPQLV1440
1441 RVGRSDVVNV AIKDLTLEEL VDKRIGERNY EIRTDPELER KFNNAVTKRR ELRGKLDSES1500
1501 GNPESPMSTE DISKLQLKIR ELSKIINELG RDRDEMREKN SVNYRNRDLD RRNAQAHILA1560
1561 VSDIICSTLS GSAHDVLATM GIKFDTVIID EACQCTELSS IIPLRYGGKR CIMVGDPNQL1620
1621 PPTVLSGAAS NFKYNQSLFV RMEKNSSPYL LDVQYRMHPS ISKFPSSEFY QGRLKDGPGM1680
1681 DILNKRPWHQ LEPLAPYKFF DIISGRQEQN AKTMSYTNME EIRVAIELVD YLFRKFDNKI1740
1741 DFTGKIGIIS PYREQMQKMR KEFARYFGGM INKSIDFNTI DGFQGQEKEI ILISCVRADD1800
1801 TKSSVGFLKD FRRMNVALTR AKTSIWVLGH QRSLAKSKLW RDLIEDAKDR SCLAYACSGF1860
1861 LDPRNNRAQS ILRKFNVPVP SEQEDDYKLP MEYITQGPDE VKSNKDTKKR RVVDEGEEAD1920
1921 KAVKKKKKEK KKEKKKSKAD DKKKNNKKAE SPSTSSGTKK KSSIFGGMSV PSAVVPKTFP1980
1981 DVDSNKKAAA VVGKKKNNKH VCFSDDVSFI PRNDEPEIKV TRSLSSVLKE KQLGLKETRT2040
2041 ISPPEISNNE DDDDEDDYTP SISDSSLMKS EANGRNNRVA SHNQNFSASI YDDPQVSQAK2100
2101 QTQVPAAITK HRSSNSVLSG GSSRILTASD YGEPNQNGQN GANRTLSQHV GNANQYSTAP2160
2161 VGTGELHETL PAHPQDSYPA EAEDPYDLNP HPQPQSSAFK GPGSGPTGTR NSSRRNASSS2220
2221 PFIPKKRKPR S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
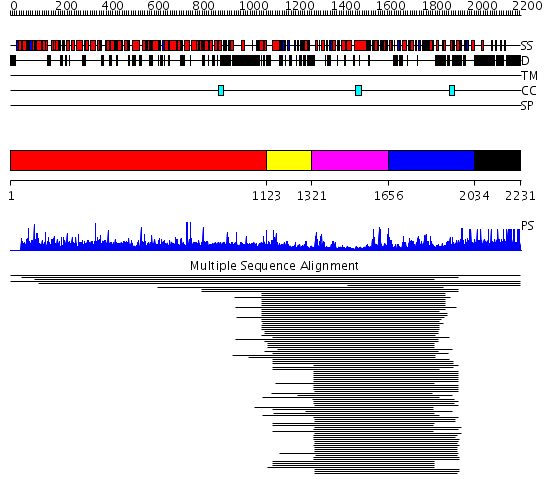
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [1123..1320] | 1.115993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1321..1655] | 97.154902 | DEXX box DNA helicase | |
| 4 | View Details | [1656..2033] | 97.154902 | DEXX box DNA helicase | |
| 5 | View Details | [2034..2231] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [841..1128] | 1.16 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | |
| 7 | View Details | [1129..1859] | 138.0 | No description for 2gjkA was found. | |
| 8 | View Details | [1860..1966] | 106.0 | STRUCTURE OF DNA HELICASE | |
| 9 | View Details | [1967..2231] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)