













| General Information: |
|
| Name(s) found: |
SAP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 897 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSQRSHHIL TRLTKIRRRP QQPLTDFTEL YSRIANETIY YLNLEEKKRY KEALQGWKAL 60
61 TTDVLFKQTL IEHNYPNTQS YTKDEVSLQN GIRELYHKSV MHLKRVKKLV REEPAPRNDM 120
121 PSSKTYTNHS SSFTRSTEPP PVFQMVPGRM MKTLRNRNAC GYKTAYSNPS LSSYGNSTSI 180
181 KRGEDAENIR VNFVPSKPLS NNASRQHKNP IEHNDPPLKK ETELYSDKYI SEPILIDLTN 240
241 DEDDHDVGIL KGHNVFDEEE SDGFEFDVSD YYDNFSEVDV EEEEEEKEER RRIKTLEAIQ 300
301 QQMSDLSVTS STSSNKSVSS SENVPGSCIQ SLPTTAPALP SLPPPPLLNV DRASSTGALK 360
361 PHSLETSTTM DSSKIRNPQI SKLMKNNHVP YLKGTKSTPT LITKSTPTFI TRSKSNTKPI 420
421 IKSNASSPTS SLTVPNSVIQ KPKTAAMAAK RVLNSKKVAS NPALNTTKKS HPILKSKTAK 480
481 VPNSSSKKTS SHPSRPVSNS KPYSHGASQN KKPSKNQTTS MSKTNRKIPA QKKIGSPKIE 540
541 DVGTEDATEH ATSLNEQREE PEIDKKVLRE ILEDEIIDSL QGVDRQAAKQ IFAEIVVHGD 600
601 EVHWDDIAGL ESAKYSLKEA VVYPFLRPDL FRGLREPVRG MLLFGPPGTG KTMLARAVAT 660
661 ESHSTFFSIS ASSLTSKYLG ESEKLVRALF AIAKKLSPSI IFVDEIDSIM GSRNNENENE 720
721 SSRRIKNEFL VQWSSLSSAA AGSNKSNTNN SDTNGDEDDT RVLVLAATNL PWSIDEAARR 780
781 RFVRRQYIPL PEDQTRHVQF KKLLSHQKHT LTESDFDELV KITEGYSGSD ITSLAKDAAM 840
841 GPLRDLGDKL LETEREMIRP IGLVDFKNSL VYIKPSVSQD GLVKYEKWAS QFGSSGS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
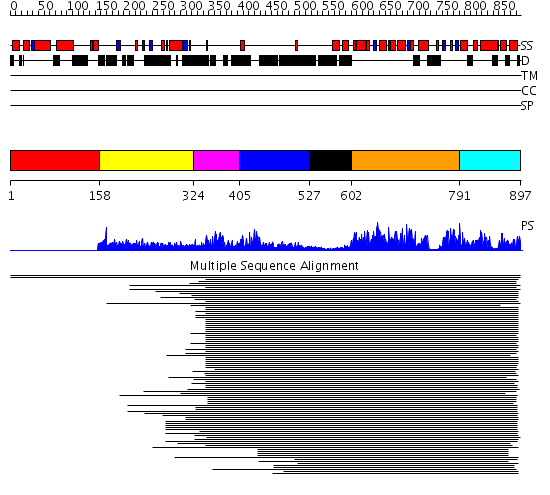
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [158..323] | 1.034991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [324..404] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [405..526] | 438.616428 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [527..601] | 438.616428 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 6 | View Details | [602..790] | 438.616428 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 7 | View Details | [791..897] | 438.616428 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)