













| General Information: |
|
| Name(s) found: |
ROG1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 685 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLTPTNEIL FHYKSSVKVG ELERYVITYH LYDGEEIPPD LNLNSLWLKV RNMNPLSYRA 60
61 AYLMGPFMLY CDVKTAQYHH SQKIVASVDY PKFEPNVQTQ QDFVAELSVH NIRQKYVWIA 120
121 DVMSQILFTT NTNVTYEVTI GTSKESVENP HDLPSHLGSY SPKLTVNRLT TLDLWNLPVQ 180
181 ITTPQKKKHL VVLTHGLHSN VSTDLVYIME QIYKAQKNYP HEQIVVKGYR GNVCQTEKGV 240
241 KYLGTRLAEY IIQDLYDESI RKISFVGHSL GGLIQAFAIA YIYEVYPWFF KKVNPINFIT 300
301 LASPLLGIVT DNPAYIKVLL SFGVIGKTGQ DLGLENDVEV GKPLLYLLSG LPLIEILRRF 360
361 KRRTVYANAI NDGIVPLYTA SLLFLDYNDI LEQLQKLKEN SKKSPLINDA STPVNQDFFN 420
421 KTFISPLTKM LSILAPQKFP TENGSEIPKV SFFESASSIL LPPLPERAYI MDPDSRDPVI 480
481 IHDKIYNEDD IPQSEFDIED GFFGKKNILL QAFFAGKKER AKYRNLEETI ARRWHEGMAW 540
541 RKVVVALKPD AHNNIIVRRK FANAYGWPVI DHLIDVHFNG DDDDDNDEND DINSTQVVEP 600
601 IQSVTEGKKK YRKAENIPQE YGWLNKVETN GVFDEGPTGM ISTVGEIVEA LAKRGFSAVI 660
661 DRRNASEDPN DEVLRFEEMN SDLVQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
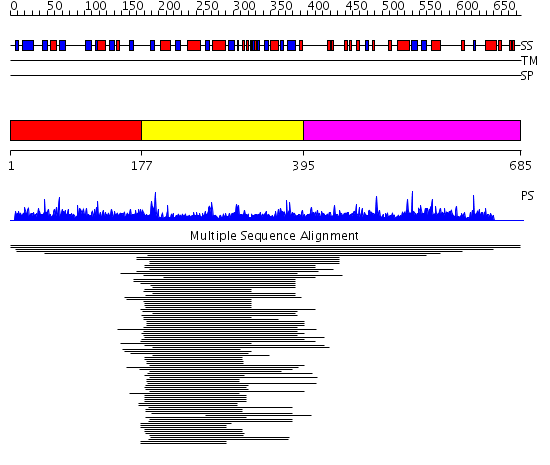
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [177..394] | 32.045757 | Lipase | |
| 3 | View Details | [395..685] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [499..584] | 1.025979 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [585..685] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)