













| General Information: |
|
| Name(s) found: |
RNA14_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 677 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSTTPDLL YPSADKVAEP SDNIHGDELR LRERIKDNPT NILSYFQLIQ YLETQESYAK 60
61 VREVYEQFHN TFPFYSPAWT LQLKGELARD EFETVEKILA QCLSGKLENN DLSLWSTYLD 120
121 YIRRKNNLIT GGQEARAVIV KAFQLVMQKC AIFEPKSSSF WNEYLNFLEQ WKPFNKWEEQ 180
181 QRIDMLREFY KKMLCVPFDN LEKMWNRYTQ WEQEINSLTA RKFIGELSAE YMKARSLYQE 240
241 WLNVTNGLKR ASPINLRTAN KKNIPQPGTS DSNIQQLQIW LNWIKWEREN KLMLSEDMLS 300
301 QRISYVYKQG IQYMIFSAEM WYDYSMYISE NSDRQNILYT ALLANPDSPS LTFKLSECYE 360
361 LDNDSESVSN CFDKCTQTLL SQYKKIASDV NSGEDNNTEY EQELLYKQRE KLTFVFCVYM 420
421 NTMKRISGLS AARTVFGKCR KLKRILTHDV YVENAYLEFQ NQNDYKTAFK VLELGLKYFQ 480
481 NDGVYINKYL DFLIFLNKDS QIKTLFETSV EKVQDLTQLK EIYKKMISYE SKFGNLNNVY 540
541 SLEKRFFERF PQENLIEVFT SRYQIQNSNL IKKLELTYMY NEEEDSYFSS GNGDGHHGSY 600
601 NMSSSDRKRL MEETGNNGNF SNKKFKRDSE LPTEVLDLLS VIPKRQYFNT NLLDAQKLVN 660
661 FLNDQVEIPT VESTKSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
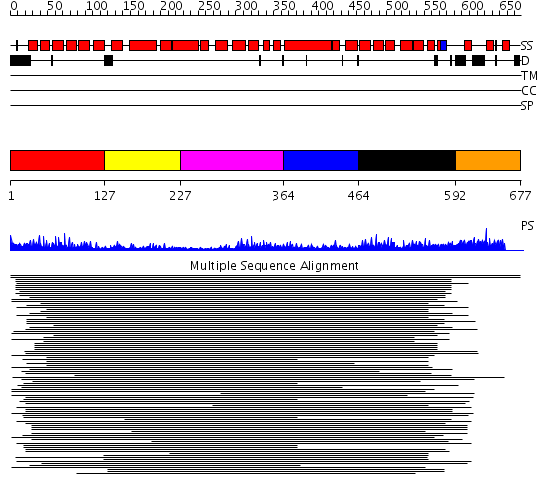
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 2.69897 | Transcription factor MalT domain III | |
| 2 | View Details | [127..226] | 2.69897 | Transcription factor MalT domain III | |
| 3 | View Details | [227..363] | 17.39794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [364..463] | 17.39794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [464..591] | 17.39794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [592..677] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)