













| General Information: |
|
| Name(s) found: |
PLDZ2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1046 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
|
| Biological Process: |
root development
[IGI]
galactolipid biosynthetic process [IMP] phospholipid catabolic process [IMP] cellular response to phosphate starvation [IEP] cellular response to nitrogen starvation [IEP] response to auxin stimulus [IMP] |
| Molecular Function: |
phospholipase D activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTDKLLLPN GVKSDGVIRM TRADAAAAAA SSSLGGGSQI FDELPKAAIV SVSRPDTTDF 60
61 SPLLLSYTLE LQYKQFKWTL QKKASQVLYL HFALKKRLII EELHDKQEQV REWLHSLGIF 120
121 DMQGSVVQDD EEPDDGALPL HYTEDSIKNR NVPSRAALPI IRPTIGRSET VVDRGRTAMQ 180
181 GYLSLFLGNL DIVNSKEVCK FLEVSRLSFA REYGSKMKEG YVTVKHLRDV PGSDGVRCCL 240
241 PTHCLGFFGT SWTKVWAVLK PGFLALLEDP FSGKLLDIMV FDTLGLQGTK ESSEQPRLAE 300
301 QVKEHNPLRF GFKVTSGDRT VRLRTTSSRK VKEWVKAVDE AGCYSPHRFG SFAPPRGLTS 360
361 DGSQAQWFVD GHTAFEAIAF AIQNATSEIF MTGWWLCPEL YLKRPFEDHP SLRLDALLET 420
421 KAKQGVKIYI LLYKEVQIAL KINSLYSKKR LQNIHKNVKV LRYPDHLSSG IYLWSHHEKI 480
481 VIVDYQVCFI GGLDLCFGRY DTAEHKIGDC PPYIWPGKDY YNPRESEPNS WEETMKDELD 540
541 RRKYPRMPWH DVHCALWGPP CRDVARHFVQ RWNHSKRNKA PNEQTIPLLM PHHHMVLPHY 600
601 LGTREIDIIA AAKPEEDPDK PVVLARHDSF SSASPPQEIP LLLPQETDAD FAGRGDLKLD 660
661 SGARQDPGET SEESDLDEAV NDWWWQIGKQ SDCRCQIIRS VSQWSAGTSQ PEDSIHRAYC 720
721 SLIQNAEHFI YIENQFFISG LEKEDTILNR VLEALYRRIL KAHEENKCFR VVIVIPLLPG 780
781 FQGGIDDFGA ATVRALMHWQ YRTISREGTS ILDNLNALLG PKTQDYISFY GLRSYGRLFE 840
841 DGPIATSQIY VHSKLMIVDD RIAVIGSSNI NDRSLLGSRD SEIGVVIEDK EFVESSMNGM 900
901 KWMAGKFSYS LRCSLWSEHL GLHAGEIQKI EDPIKDATYK DLWMATAKKN TDIYNQVFSC 960
961 IPNEHIRSRA ALRHNMALCK DKLGHTTIDL GIAPERLESC GSDSWEILKE TRGNLVCFPL1020
1021 QFMCDQEDLR PGFNESEFYT APQVFH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
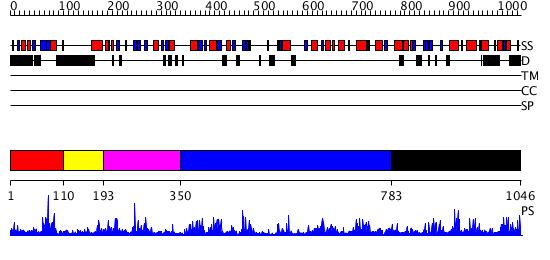
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 2.049991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [110..192] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [193..349] | 5.677781 | No description for PF00169.21 was found. No confident structure predictions are available. | |
| 4 | View Details | [350..782] | 21.30103 | Tungstate-inhibited phospholipase D from Streptomyces sp. strain PMF. | |
| 5 | View Details | [783..1046] | 1.231961 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)