













| General Information: |
|
| Name(s) found: |
REG1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1014 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTNLANYFA GKKDIENEHV NRNASHESNS KSDVKISGND NDNDEDMGPS VSMAVQAKND 60
61 DDFHKSTFNL KRTRSMGLLD EYIDPTKKLL GRSDDLYDND NEYYDNSSNN SSSNSSDDDY 120
121 DDGYQEHSTS VSPPPADNDS YLIPQDDNDV VVEPERHVDY LSHEWKESEI SNSWKYIILK 180
181 KKKRDVDLVN AARLENASWR TWAKARNNLK TVSPEVVNWS KDSDVTWLYG PIVRDSEGNA 240
241 QSEEEHDLER GYGSDDENSK RISMPTKNSK SIAAAPKPIL KKRTVTEIIE DNALWKLNEA 300
301 RKHMTEMKHA SVIMDPNGNK NVHDDFDALA AQVNAQYYHY PKESNSSVSL KSQHSDKKDN 360
361 STIPNPVGEN SNGGGDKGEE DLHLKSALHV QNNRSTAQSN KSILENSTND RKANLDQNLN 420
421 SPDNNRFPSS TSSSNRDNEN NSMGLSSILT SNPSEKSNKP TKNRHIHFND RVEQCMALRY 480
481 PASQSEDDES DDENKQYVDV NNNANVTTIN NNRTPLLAIQ HKSIPINSAT EHLNKNTSDD 540
541 DTSSQSSSSS HSDDEEHGGL YINARFSRRS DSGVHSPITD NSSVASSTTS RAHVRPIIKL 600
601 LPDTTLNYGS DEESDNGEFN GYGNAVSHNV NTSRGYDYIY DYNSVYTGDT SSFLPVDSCD 660
661 IVDVPEGMDL QTAIADDNAS NYEFNNAVES KEKHVPQLHK ASANNTTRQH GSHMLLYDDD 720
721 NYSSSSDSEQ QFIEDSQYNS SDDEEEEDDD DQEVDDNHDE GLSLRRTLSL GKSGSTNSLY 780
781 DLAQPSLSSA TPQQKNPTNF TGGKTDVDKD AQLAVRPYPL KRNSSSGNFI FNSDSEEESS 840
841 SEEEQRPLPA NSQLVNRSVL KGSVTPANIS SQKKKALPKQ PKASDSSQSF RIVNNTPSPA 900
901 EVGASDVAIE GYFSPRNESI KSVVSGGNMM DHQDHSEMDT LAKGFENCHI NNASKLKDKK 960
961 VDSVQTTRKE ASLTDSSNES LHKVVQNARG MASKYLHSWK KSDVKPQENG NDSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
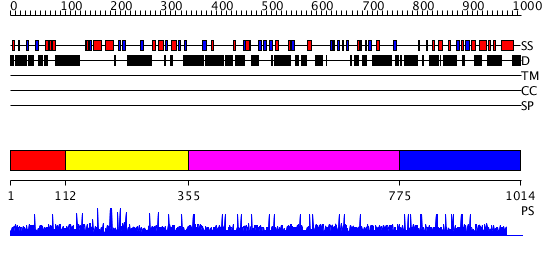
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [112..354] | 2.004977 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [355..774] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [775..1014] | 1.002892 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)