













| General Information: |
|
| Name(s) found: |
PWP2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 923 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA 60
61 IDLNKQGTLL ISIDEDGRAI LVNFKARNVL HHFNFKEKCS AVKFSPDGRL FALASGRFLQ 120
121 IWKTPDVNKD RQFAPFVRHR VHAGHFQDIT SLTWSQDSRF ILTTSKDLSA KIWSVDSEEK 180
181 NLAATTFNGH RDYVMGAFFS HDQEKIYTVS KDGAVFVWEF TKRPSDDDDN ESEDDDKQEE 240
241 VDISKYSWRI TKKHFFYANQ AKVKCVTFHP ATRLLAVGFT SGEFRLYDLP DFTLIQQLSM 300
301 GQNPVNTVSV NQTGEWLAFG SSKLGQLLVY EWQSESYILK QQGHFDSTNS LAYSPDGSRV 360
361 VTASEDGKIK VWDITSGFCL ATFEEHTSSV TAVQFAKRGQ VMFSSSLDGT VRAWDLIRYR 420
421 NFRTFTGTER IQFNCLAVDP SGEVVCAGSL DNFDIHVWSV QTGQLLDALS GHEGPVSCLS 480
481 FSQENSVLAS ASWDKTIRIW SIFGRSQQVE PIEVYSDVLA LSMRPDGKEV AVSTLKGQIS 540
541 IFNIEDAKQV GNIDCRKDII SGRFNQDRFT AKNSERSKFF TTIHYSFDGM AIVAGGNNNS 600
601 ICLYDVPNEV LLKRFIVSRN MALNGTLEFL NSKKMTEAGS LDLIDDAGEN SDLEDRIDNS 660
661 LPGSQRGGDL STRKMRPEVR VTSVQFSPTA NAFAAASTEG LLIYSTNDTI LFDPFDLDVD 720
721 VTPHSTVEAL REKQFLNALV MAFRLNEEYL INKVYEAIPI KEIPLVASNI PAIYLPRILK 780
781 FIGDFAIESQ HIEFNLIWIK ALLSASGGYI NEHKYLFSTA MRSIQRFIVR VAKEVVNTTT 840
841 DNKYTYRFLV STDGSMEDGA ADDDEVLLKD DADEDNEENE ENDVVMESDD EEGWIGFNGK 900
901 DNKLPLSNEN DSSDEEENEK ELP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
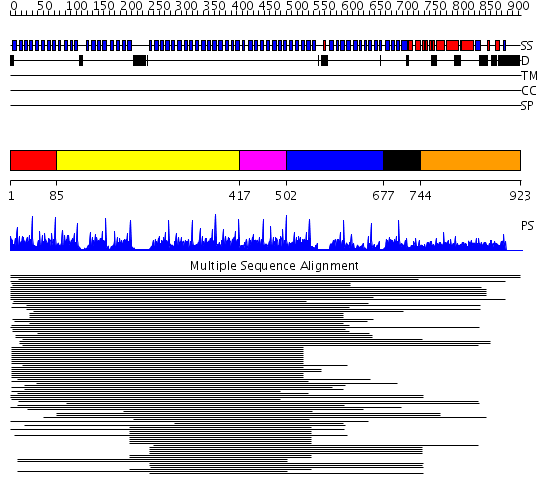
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 1.441995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [85..416] | 131.274545 | Groucho/tle1, C-terminal domain | |
| 3 | View Details | [417..501] | 252.69897 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 4 | View Details | [502..676] | 202.9897 | Tup1, C-terminal domain | |
| 5 | View Details | [677..743] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [744..923] | 2.054975 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)