













| General Information: |
|
| Name(s) found: |
PRP16_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1071 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGHSGREERI KDIFKELTSK ELTPGLLLTL QKLAQKPNTN LEQFIASCKA LTKLSSNNPI 60
61 IFNELLELLK NKSEEDSTGP KKIAPSINKR KKFKIQLDLD DNEDELDSPV QKKPAPTRTL 120
121 FKRIDKLKAK QLRQYSPTVK DPSPNSEQQT QNGHAETKDY EPTRSEVVEE DREWYDNDDD 180
181 YGNLVPEPLS ELPEEAKLLP VIRNIDNDDA LRNTVQLYPI PLKQRMEWIP PFLSKFALEN 240
241 KVPTSIIIGS ISETSSQVSA LSMVNPFRNP DSEFSANAKR GSKLVALRRI NMEHIQQSRD 300
301 NTTVLNTAMG EVLGLENNNK AKDKSNQKIC DDTALFTPSK DDIKHTKEQL PVFRCRSQLL 360
361 SLIRENQVVV IIGETGSGKT TQLAQYLYEE GYANDRGKSI VVTQPRRVAA ISVAKRVAME 420
421 MQVPLGKEVG YSIRFEDVTD SECTKLKFVT DGILLRETLL DDTLDKYSCV IIDEAHERSL 480
481 NTDILLGFFK ILLARRRDLK LIITSATMNA KKFSAFFGNA PQFTIPGRTF PVQTIYTSNP 540
541 VQDYVEAAVS QAVKIHLAND CSSGDILIFM TGQEDIETTF DTLQEKFLQV YSKKFGTANF 600
601 EEINDIEILP IYSALPADLQ FKIFQDLHGT KRKIIIATNI AETSLTIKGI RYVIDCGYSK 660
661 LKVYNPKIGL DSLVITPISK ANADQRSGRA GRTAPGTAYR LYTEDTFKED MYLQTIPEIQ 720
721 RTNLSNTLLL LKSLDVTDEL SKFPFIDKPP LQTFLSSLYE LWFIGAIDTS GQLTPLGLQM 780
781 AKFPLQPSLS KILLIAVRNG CSDEMLTIVS MLSVPQVFYR PKERQKEADI ARNKFFIAKS 840
841 DHLTLLNVFE QWRANNFSSH WCNKHFVQYK SLVRARDIRD QLLTILKSQK IPVISSGKDW 900
901 DIIKKCICSG FAHQAAKITG LRNYVHLKTG VSVQLHPTSA LHGLGDLPPY VVYHELLMTS 960
961 KEYICCVTSV DPFWLMEYGG LLYDIKRIKN DQEATTTGLF GEHYEHTLDK VEDDIDINIR1020
1021 RCKDMRDSVI QELKMTDNSN KEDKKQKTKK QNILNGKENS MKPFKRRKPF F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
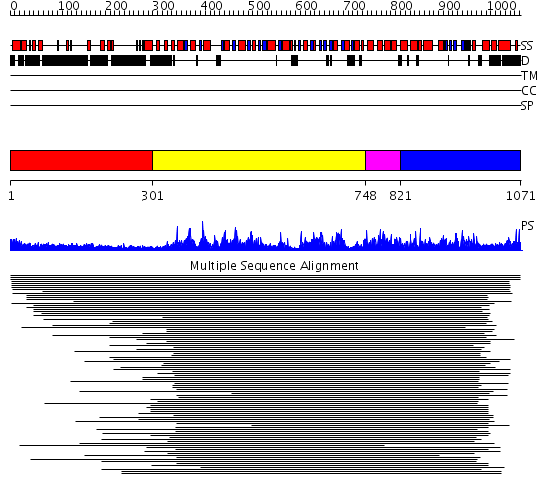
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..300] | 17.445997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [301..747] | 62.69897 | Initiation factor 4a | |
| 3 | View Details | [748..820] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [821..1071] | 3.165976 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [899..1071] | 2.562986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)