













| General Information: |
|
| Name(s) found: |
PDS5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1277 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKGAVTKLK FNSPIISTSD QLISTNELLD RLKALHEELA SLDQDNTDLT GLDKYRDALV 60
61 SRKLLKHKDV GIRAFTACCL SDILRLYAPD APYTDAQLTD IFKLVLSQFE QLGDQENGYH 120
121 IQQTYLITKL LEYRSIVLLA DLPSSNNLLI ELFHIFYDPN KSFPARLFNV IGGILGEVIS 180
181 EFDSVPLEVL RLIFNKFLTY NPNEIPEGLN VTSDCGYEVS LILCDTYSNR MSRHLTKYYS 240
241 EIIHEATNDD NNSRLLTVVV KLHKLVLRLW ETVPELINAV IGFIYHELSS ENELFRKEAT 300
301 KLIGQILTSY SDLNFVSTHS DTFKAWISKI ADISPDVRVE WTESIPQIIA TREDISKELN 360
361 QALAKTFIDS DPRVRRTSVM IFNKVPVTEI WKNITNKAIY TSLLHLAREK HKEVRELCIN 420
421 TMAKFYSNSL NEIERTYQNK EIWEIIDTIP STLYNLYYIN DLNINEQVDS VIFEYLLPFE 480
481 PDNDKRVHRL LTVLSHFDKK AFTSFFAFNA RQIKISFAIS KYIDFSKFLN NQESMSSSQG 540
541 PIVMNKYNQT LQWLASGLSD STKAIDALET IKQFNDERIF YLLNACVTND IPFLTFKNCY 600
601 NELVSKLQTP GLFKKYNIST GASIMPRDIA KVIQILLFRA SPIIYNVSNI SVLLNLSNNS 660
661 DAKQLDLKRR ILDDISKVNP TLFKDQIRTL KTIIKDLDDP DAEKNDNLSL EEALKTLYKA 720
721 SKTLKDQVDF DDTFFFTKLY DFAVESKPEI TKYATKLIAL SPKAEETLKK IKIRILPLDL 780
781 QKDKYFTSHI IVLMEIFKKF PHVLNDDSTD IISYLIKEVL LSNQVVGDSK KEIDWVEDSL 840
841 LSDTKYSAIG NKVFTLKLFT NKLRSIAPDV PRDELAESFT EKTMKLFFYL IASGGELISE 900
901 FNKEFYPTPS NYQTKLRCVA GIQVLKLARI SNLNNFIKPS DIIKLINLVE DESLPVRKTF 960
961 LEQLKDYVAN ELISIKFLPL VFFTAYEPDV ELKTTTKIWI NFTFGLKSFK KGTIFERALP1020
1021 RLIHAIAHHP DIVGGLDSEG DAYLNALTTA IDYLLFYFDS IAAQENFSLL YYLSERVKNY1080
1081 QDKLVEDEID EEEGPQKEEA PKKHRPYGQK MYIIGELSQM ILLNLKEKKN WQHSAYPGKL1140
1141 NLPSDLFKPF ATVQEAQLSF KTYIPESLTE KIQNNIKAKI GRILHTSQTQ RQRLQKRLLA1200
1201 HENNESQKKK KKVHHARSQA DDEEGDGDRE SDSDDDSYSP SNKNETKKGH ENIVMKKLRV1260
1261 RKEVDYKDDE DDDIEMT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
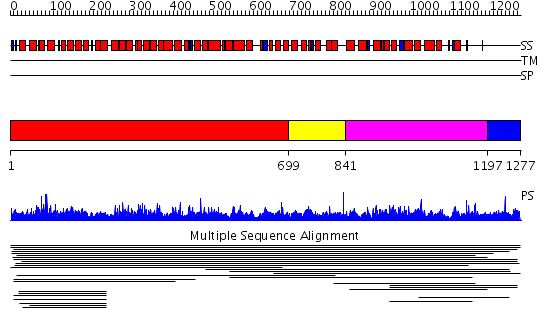
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..698] | 1.016001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [699..840] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [841..1196] | 2.017915 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1197..1277] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)