













| General Information: |
|
| Name(s) found: |
NUP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1076 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSNTSSVMS SPRVEKRSFS STLKSFFTNP NKKRPSSKKV FSSNLSYANH LEESDVEDTL 60
61 HVNKRKRVSG TSQHSDSLTQ NNNNAPIIIY GTENTERPPL LPILPIQRLR LLREKQRVRN 120
121 MRELGLIQST EFPSITSSVI LGSQSKSDEG GSYLCTSSTP SPIKNGSCTR QLAGKSGEDT 180
181 NVGLPILKSL KNRSNRKRFH SQSKGTVWSA NFEYDLSEYD AIQKKDNKDK EGNAGGDQKT 240
241 SENRNNIKSS ISNGNLATGP NLTSEIEDLR ADINSNRLSN PQKNLLLKGP ASTVAKTAPI 300
301 QESFVPNSER SGTPTLKKNI EPKKDKESIV LPTVGFDFIK DNETPSKKTS PKATSSAGAV 360
361 FKSSVEMGKT DKSTKTAEAP TLSFNFSQKA NKTKAVDNTV PSTTLFNFGG KSDTVTSASQ 420
421 PFKFGKTSEK SENHTESDAP PKSTAPIFSF GKQEENGDEG DDENEPKRKR RLPVSEDTNT 480
481 KPLFDFGKTG DQKETKKGES EKDASGKPSF VFGASDKQAE GTPLFTFGKK ADVTSNIDSS 540
541 AQFTFGKAAT AKETHTKPSE TPATIVKKPT FTFGQSTSEN KISEGSAKPT FSFSKSEEER 600
601 KSSPISNEAA KPSFSFPGKP VDVQAPTDDK TLKPTFSFTE PAQKDSSVVS EPKKPSFTFA 660
661 SSKTSQPKPL FSFGKSDAAK EPPGSNTSFS FTKPPANETD KRPTPPSFTF GGSTTNNTTT 720
721 TSTKPSFSFG APESMKSTAS TAAANTEKLS NGFSFTKFNH NKEKSNSPTS FFDGSASSTP 780
781 IPVLGKPTDA TGNTTSKSAF SFGTANTNGT NASANSTSFS FNAPATGNGT TTTSNTSGTN 840
841 IAGTFNVGKP DQSIASGNTN GAGSAFGFSS SGTAATGAAS NQSSFNFGNN GAGGLNPFTS 900
901 ATSSTNANAG LFNKPPSTNA QNVNVPSAFN FTGNNSTPGG GSVFNMNGNT NANTVFAGSN 960
961 NQPHQSQTPS FNTNSSFTPS TVPNINFSGL NGGITNTATN ALRPSDIFGA NAASGSNSNV1020
1021 TNPSSIFGGA GGVPTTSFGQ PQSAPNQMGM GTNNGMSMGG GVMANRKIAR MRHSKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
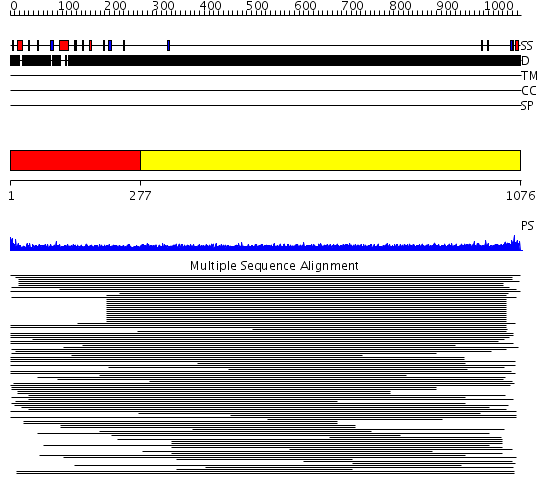
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..276] | 1.150997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [277..1076] | 10.232997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [389..632] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [633..746] | 2.214988 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [747..892] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [893..1076] | 2.19299 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)