













| General Information: |
|
| Name(s) found: |
NU170_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1502 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFQSFFHNNG PAAAGETFSD SRSYPLTNHQ EVPRNGLNEL ASSATKAQQQ PTHILNSYPI 60
61 TGSNPLMRAS AMGATSGSIN PNMSNMNEHI RVSGMGTSKP LDLAGKYIDH LQHKDSNTPV 120
121 LDERSYYNSG VDYNFSREKN GLGAFTPFEK QDVFNIPDEI LHEFSTSQTK TDMGIFPELN 180
181 RCWITIDNKL ILWNINNDNE YQVVDDMKHT IQKVALVRPK PNTFVPAVKH LLLISTTMEL 240
241 FMFAISLDKA TNELSVFNTH LSVPVQGIDV IDIVSHERSG RIFFAGQASG LNIWELHYSG 300
301 SDDWFNSKCS KVCLTKSALL SLLPTNMLSQ IPGVDFIQAL FEDNSNGNGG FSQETITQLT 360
361 IDQQRGIIYS LSSKSTIRAY VITEKSLEGP MSIEPAYISR IIGTTTARAA PILGPKYLKI 420
421 VKISSVAPEE NNNLFLVALT VGGVRLYFNG SMGRFNIEAL RLESIKFPPS SVTPEVIQQE 480
481 LLHQQQEQAK RSFPFFSNLM SSEPVLLKFQ KKSSVLLETT KASTIISPGI FFSAVIKSSQ 540
541 QTHQQEKKEN SSVTGTTATA GSKTVKQQPV TLQHKLFVSV PDYGILKTHG KYVENATFLE 600
601 TAGPVQQIIP LSGLFNATTK PQGFANEFAT QYTSETLRVA VLTSTSIEIY KYRTPDEIFE 660
661 DLIDNPLPFV LNYGAAEACS TALFVTCKSN KSEKLRSNAL TFLTMGIPGV VDIKPVYNRY 720
721 SVSTVSSLLS KPTLSTATTN LQQSITGFSK PSPANKEDFD LDDVILSPRF YGIALLITRL 780
781 LRDIWGRHVF MTFTDNRVTS HAFISSSDPI TPSINNLKSD EISQNRNIIS KVSISKDCIE 840
841 YYLSSINILN EFFITYGDSI SQISAPYVLA NNSNGRVIDK TEEVANQAES IAINAMIKMV 900
901 QSIKEGLSFL NVLYEESEVE GFDNQYLGFK DIISFVSLDV QKDLVKLDFK DLFAPNDKTK 960
961 SLIREILLSI INRNITKGAS IEYTATALQE RCGSFCSASD ILGFRAIEHL RRAKEIGLRN1020
1021 YDSLNYHLKN ATALLEQIVD DLSIEKLKEA VSMMLSVNYY PKSIEFLLNI ANSMDKGKLA1080
1081 CQYVANGFLE NDDRKQYYDK RILVYDLVFD TLIKVDELAE KKQSSKTQNQ ISISNDDEVK1140
1141 LRQKSYEAAL KYNDRLFHYH MYDWLVSQNR EEKLLDIETP FILPYLMEKA GSSLKISNIL1200
1201 WVYYSRRSKF FESAEILYRL ATSNFDITLF ERIEFLSRAN GFCNSVSPLS QKQRIVQLAS1260
1261 RIQDACEVAG IQGDILSLVY TDARIDSAIK DELIKTLDGK ILSTSELFND FAVPLSYHEI1320
1321 ALFIFKIADF RDHEVIMAKW DELFQSLRME FNNTGKKEDS MNFINLLSNV LIKIGKNVQD1380
1381 SEFIFPIFEL FPIVCNFFYE TLPKEHIVSG SIVSIFITAG VSFNKMYYIL KELIETSDSD1440
1441 NSVFNKEMTW LIHEWYKSDR KFRDIISYND IIHLKEYKID NDPIEKYVKN SGNNLGICFY1500
1501 KE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
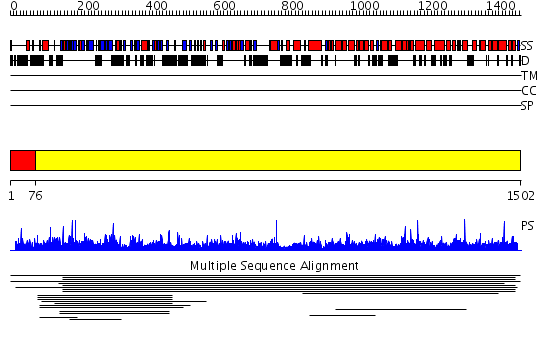
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..1502] | 1000.0 | Non-repetitive/WGA-negative nucleoporin No confident structure predictions are available. | |
| 3 | View Details | [477..712] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [713..871] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [872..954] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [955..1016] | 1.037995 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1017..1152] | 1.039976 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1153..1349] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1350..1502] | 2.037993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)