













| General Information: |
|
| Name(s) found: |
NU159_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1460 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLKDEVPT ETSEDFGFKF LGQKQILPSF NEKLPFASLQ NLDISNSKSL FVAASGSKAV 60
61 VGELQLLRDH ITSDSTPLTF KWEKEIPDVI FVCFHGDQVL VSTRNALYSL DLEELSEFRT 120
121 VTSFEKPVFQ LKNVNNTLVI LNSVNDLSAL DLRTKSTKQL AQNVTSFDVT NSQLAVLLKD 180
181 RSFQSFAWRN GEMEKQFEFS LPSELEELPV EEYSPLSVTI LSPQDFLAVF GNVISETDDE 240
241 VSYDQKMYII KHIDGSASFQ ETFDITPPFG QIVRFPYMYK VTLSGLIEPD ANVNVLASSC 300
301 SSEVSIWDSK QVIEPSQDSE RAVLPISEET DKDTNPIGVA VDVVTSGTIL EPCSGVDTIE 360
361 RLPLVYILNN EGSLQIVGLF HVAAIKSGHY SINLESLEHE KSLSPTSEKI PIAGQEQEEK 420
421 KKNNESSKAL SENPFTSANT SGFTFLKTQP AAANSLQSQS SSTFGAPSFG SSAFKIDLPS 480
481 VSSTSTGVAS SEQDATDPAS AKPVFGKPAF GAIAKEPSTS EYAFGKPSFG APSFGSGKSS 540
541 VESPASGSAF GKPSFGTPSF GSGNSSVEPP ASGSAFGKPS FGTPSFGSGN SSAEPPASGS 600
601 AFGKPSFGTS AFGTASSNET NSGSIFGKAA FGSSSFAPAN NELFGSNFTI SKPTVDSPKE 660
661 VDSTSPFPSS GDQSEDESKS DVDSSSTPFG TKPNTSTKPK TNAFDFGSSS FGSGFSKALE 720
721 SVGSDTTFKF GTQASPFSSQ LGNKSPFSSF TKDDTENGSL SKGSTSEIND DNEEHESNGP 780
781 NVSGNDLTDS TVEQTSSTRL PETPSDEDGE VVEEEAQKSP IGKLTETIKK SANIDMAGLK 840
841 NPVFGNHVKA KSESPFSAFA TNITKPSSTT PAFSFGNSTM NKSNTSTVSP MEEADTKETS 900
901 EKGPITLKSV ENPFLPAKEE RTGESSKKDH NDDPKDGYVS GSEISVRTSE SAFDTTANEE 960
961 IPKSQDVNNH EKSETDPKYS QHAVVDHDNK SKEMNETSKN NERSGQPNHG VQGDGIALKK1020
1021 DNEKENFDSN MAIKQFEDHQ SSEEDASEKD SRQSSEVKES DDNMSLNSDR DESISESYDK1080
1081 LEDINTDELP HGGEAFKARE VSASADFDVQ TSLEDNYAES GIQTDLSESS KENEVQTDAI1140
1141 PVKHNSTQTV KKEAVDNGLQ TEPVETCNFS VQTFEGDENY LAEQCKPKQL KEYYTSAKVS1200
1201 NIPFVSQNST LRLIESTFQT VEAEFTVLME NIRNMDTFFT DQSSIPLVKR TVRSINNLYT1260
1261 WRIPEAEILL NIQNNIKCEQ MQITNANIQD LKEKVTDYVR KDIAQITEDV ANAKEEYLFL1320
1321 MHFDDASSGY VKDLSTHQFR MQKTLRQKLF DVSAKINHTE ELLNILKLFT VKNKRLDDNP1380
1381 LVAKLAKESL ARDGLLKEIK LLREQVSRLQ LEEKGKKASS FDASSSITKD MKGFKVVEVG1440
1441 LAMNTKKQIG DFFKNLNMAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
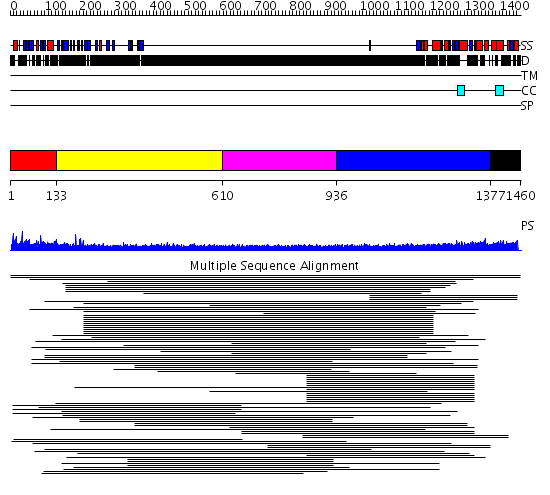
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [133..609] | 8.207997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [610..935] | 1.212997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [936..1376] | 10.217996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1377..1460] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)