













| General Information: |
|
| Name(s) found: |
NU116_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1113 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGVSRGAFP SATTQPFGST GSTFGGQQQQ QQPVANTSAF GLSQQTNTTQ APAFGNFGNQ 60
61 TSNSPFGMSG STTANGTPFG QSQLTNNNAS GSIFGGMGNN TALSAGSASV VPNSTAGTSI 120
121 KPFTTFEEKD PTTGVINVFQ SITCMPEYRN FSFEELRFQD YQAGRKFGTS QNGTGTTFNN 180
181 PQGTTNTGFG IMGNNNSTTS ATTGGLFGQK PATGMFGTGT GSGGGFGSGA TNSTGLFGSS 240
241 TNLSGNSAFG ANKPATSGGL FGNTTNNPTN GTNNTGLFGQ QNSNTNGGLF GQQQNSFGAN 300
301 NVSNGGAFGQ VNRGAFPQQQ TQQGSGGIFG QSNANANGGA FGQQQGTGAL FGAKPASGGL 360
361 FGQSAGSKAF GMNTNPTGTT GGLFGQTNQQ QSGGGLFGQQ QNSNAGGLFG QNNQSQNQSG 420
421 LFGQQNSSNA FGQPQQQGGL FGSKPAGGLF GQQQGASTFA SGNAQNNSIF GQNNQQQQST 480
481 GGLFGQQNNQ SQSQPGGLFG QTNQNNNQPF GQNGLQQPQQ NNSLFGAKPT GFGNTSLFSN 540
541 STTNQSNGIS GNNLQQQSGG LFQNKQQPAS GGLFGSKPSN TVGGGLFGNN QVANQNNPAS 600
601 TSGGLFGSKP ATGSLFGGTN STAPNASSGG IFGSNNASNT AATTNSTGLF GNKPVGAGAS 660
661 TSAGGLFGNN NNSSLNNSNG STGLFGSNNT SQSTNAGGLF QNNTSTNTSG GGLFSQPSQS 720
721 MAQSQNALQQ QQQQQRLQIQ NNNPYGTNEL FSKATVTNTV SYPIQPSATK IKADERKKAS 780
781 LTNAYKMIPK TLFTAKLKTN NSVMDKAQIK VDPKLSISID KKNNQIAISN QQEENLDESI 840
841 LKASELLFNP DKRSFKNLIN NRKMLIASEE KNNGSQNNDM NFKSKSEEQE TILGKPKMDE 900
901 KETANGGERM VLSSKNDGED SATKHHSRNM DEENKENVAD LQKQEYSEDD KKAVFADVAE 960
961 KDASFINENY YISPSLDTLS SYSLLQLRKV PHLVVGHKSY GKIEFLEPVD LAGIPLTSLG1020
1021 GVIITFEPKT CIIYANLPNR PKRGEGINVR ARITCFNCYP VDKSTRKPIK DPNHQLVKRH1080
1081 IERLKKNPNS KFESYDADSG TYVFIVNHAA EQT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
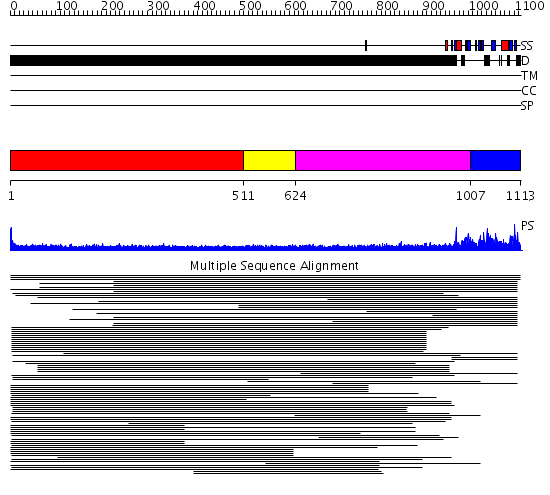
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..510] | 12.136996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [511..623] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [624..1006] | 7.152997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1007..1113] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [683..781] | 2.300996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [782..967] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [968..1113] | 64.154902 | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)