













| General Information: |
|
| Name(s) found: |
NOC3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 663 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRNRSQFR IQERTAKKRK HEDSLLEGNV FQNAPEDMDE NTIYSAKGSS WDEEEQDYEM 60
61 VPRKNRSDTS NLVEGLPIKV NGKVERKLHK AQEKPKDDDE EDEDSNDSSE DDEGPNEEQE 120
121 AEAKEDEPDT EEKILQLKED IADLVTKVME EPEENTAALG RLCKMVESKN PNTCKFSMLA 180
181 LVPVFKSIIP GYRIRPLTET EKKEKVSKEV SKLRNFEQAL VYNYKNYVGR LQSLSKTPSN 240
241 AAPIQVSLGI LATQAAKELI STASHFNFRT DIFTLLLRRI CKPRISTDPT SIQIIQTFET 300
301 LLNEDEEGSI SFEILRIFNK ILKTRNFNIE ESVLNMLLSL DVLHDYDPNT KLKGNVSAPK 360
361 LKKKDRVHLS KKQRKARKEM QQIEEEMRNA EQAVSAEERE RNQSEILKIV FTIYLNILKN 420
421 NAKTLIGSVL EGLTKFGNMA NFDLLGDFLE VMKELISDTE FDNLSSAEVR KALLCIVSAF 480
481 SLISNTQYMK VNVDLSKFVD GLYALLPYIC LDADIELSYR SLRLADPLNN EIIKPSVNVS 540
541 TKAELLLKAL DHVFFRSKSG TKERATAFTK RLYMCISHTP EKTSIAILKF IDKLMNRYPE 600
601 ISGLYSSEDR IGNGHFIMEA DNPSRSNPEA ATLWDNALLE KHYCPVVTKG LRSLSSRSKE 660
661 CSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
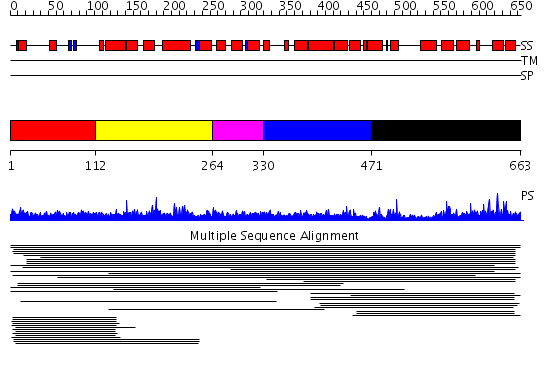
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [112..263] | 9.86 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [264..329] | 9.86 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [330..470] | 9.86 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [471..663] | 9.86 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)