













| General Information: |
|
| Name(s) found: |
NEW1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1196 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPKKFKDLN SFLDDQPKDP NLVASPFGGY FKNPAADAGS NNASKKSSYQ QQRNWKQGGN 60
61 YQQGGYQSYN SNYNNYNNYN NYNNYNNYNN YNKYNGQGYQ KSTYKQSAVT PNQSGTPTPS 120
121 ASTTSLTSLN EKLSNLELTP ISQFLSKIPE CQSITDCKNQ IKLIIEEFGK EGNSTGEKIE 180
181 EWKIVDVLSK FIKPKNPSLV RESAMLIISN IAQFFSGKPP QEAYLLPFFN VALDCISDKE 240
241 NTVKRAAQHA IDSLLNCFPM EALTCFVLPT ILDYLSSGAK WQAKMAALSV VDRIREDSAN 300
301 DLLELTFKDA VPVLTDVATD FKPELAKQGY KTLLDYVSIL DNLDLSPRYK LIVDTLQDPS 360
361 KVPESVKSLS SVTFVAEVTE PSLSLLVPIL NRSLNLSSSS QEQLRQTVIV VENLTRLVNN 420
421 RNEIESFIPL LLPGIQKVVD TASLPEVREL AEKALNVLKE DDEADKENKF SGRLTLEEGR 480
481 DFLLDHLKDI KADDSCFVKP YMNDETVIKY MSKILTVDSN VNDWKRLEDF LTAVFGGSDS 540
541 QREFVKQDFI HNLRALFYQE KERADEDEGI EIVNTDFSLA YGSRMLLNKT NLRLLKGHRY 600
601 GLCGRNGAGK STLMRAIANG QLDGFPDKDT LRTCFVEHKL QGEEGDLDLV SFIALDEELQ 660
661 STSREEIAAA LESVGFDEER RAQTVGSLSG GWKMKLELAR AMLQKADILL LDEPTNHLDV 720
721 SNVKWLEEYL LEHTDITSLI VSHDSGFLDT VCTDIIHYEN KKLAYYKGNL AAFVEQKPEA 780
781 KSYYTLTDSN AQMRFPPPGI LTGVKSNTRA VAKMTDVTFS YPGAQKPSLS HVSCSLSLSS 840
841 RVACLGPNGA GKSTLIKLLT GELVPNEGKV EKHPNLRIGY IAQHALQHVN EHKEKTANQY 900
901 LQWRYQFGDD REVLLKESRK ISEDEKEMMT KEIDIDDGRG KRAIEAIVGR QKLKKSFQYE 960
961 VKWKYWKPKY NSWVPKDVLV EHGFEKLVQK FDDHEASREG LGYRELIPSV ITKHFEDVGL1020
1021 DSEIANHTPL GSLSGGQLVK VVIAGAMWNN PHLLVLDEPT NYLDRDSLGA LAVAIRDWSG1080
1081 GVVMISHNNE FVGALCPEQW IVENGKMVQK GSAQVDQSKF EDGGNADAVG LKASNLAKPS1140
1141 VDDDDSPANI KVKQRKKRLT RNEKKLQAER RRLRYIEWLS SPKGTPKPVD TDDEED |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
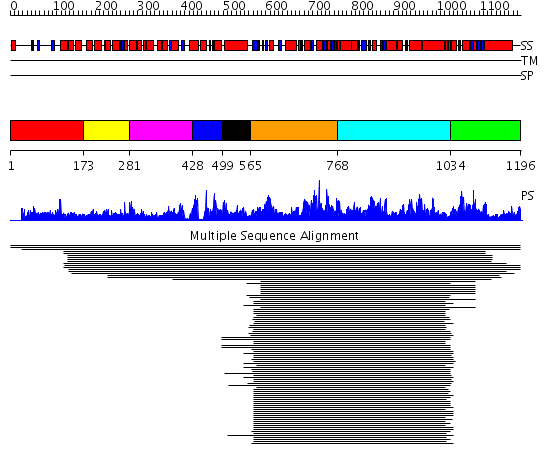
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 1.018941 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [173..280] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [281..427] | 6.09691 | Crystal structure of the P. furiosus Rad50 ATPase domain | |
| 4 | View Details | [428..498] | 6.09691 | Crystal structure of the P. furiosus Rad50 ATPase domain | |
| 5 | View Details | [499..564] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [565..767] | 118.045757 | Glucose transport protein GlcV, N-terminal domain | |
| 7 | View Details | [768..1033] | 93.679424 | MsbA | |
| 8 | View Details | [1034..1196] | 2.692976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)