













| General Information: |
|
| Name(s) found: |
MSC3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 728 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVFGFTKRDR RVPDLSRYDY YYQNHEDYNK SPQLSAAAAS AASAASPDRT NYSRSHSLVS 60
61 HAPSIPRQRS SVKSPGRRLS TSSAAPPTSR AAAKQYSQKT YSLRSQRSGE YHLHPPGYTT 120
121 NGSRMNSMTS GANVRRNYGK NKSTAGNNND SRANSITVKT TQVTDPSGRT QSITKKTIRK 180
181 INGYEYVETT TTTKNLVPLG DSQRHFDEFS ENYMLQDDDI LEEQASDNIH DIIEENETDN 240
241 EKPYSPVSES HLQDDSELNV EKPDFPLGSY FHHKYSTDVM PLEEESSLSN FSDALDYIPP 300
301 THQTSSKYIH NKRKQASTTR RKKRPPAVKN AEAEAKKPLT EAEMYLKALE VAKRNVYHTD 360
361 AASDNASAPL GSNKSRKSRM GQKMTLRSSS DSPTATANLV KSNVEVQPKR FTSSFFSRNT 420
421 KSAPHEVHNH SVSTHFKSNK AVDPVPEPKS ANTGLTDKEM YDQALKIAQA RYYNSHGIQP 480
481 EAVDNSTTAA KPRQVGVSHL GSTGSIPPNE QHYLGDSEIP VQSEVHEYEP IPLQKTKTTG 540
541 SSKNKFKTMF DKVLQFSQEN YGYQHKKEQG EQTPVTRNAE ESFPAASISE GVTTAKPSSN 600
601 EGVMTNPVVT DSPSPLQQQI DSTTASSNGQ SQGNVPTSAV ASTTRTRSPE LQDNLKSSSS 660
661 LLQDQTPQRQ EDATDPTTSS TNELSAAEPT MVTSTHATKT IQAQTQDPPT KHKKSSFFTK 720
721 LFKKKSSR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
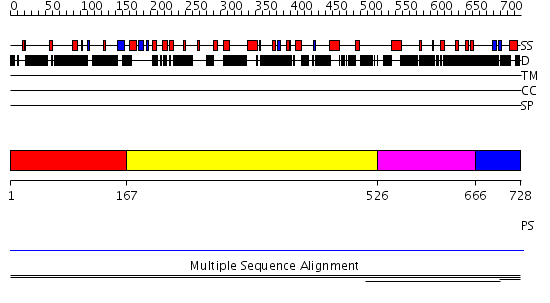
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 11.14 | Fibrinogen | |
| 2 | View Details | [167..525] | 10.93 | Hybrid domain of integrin beta; Integrin beta A domain; Integrin beta tail domain; Integrin beta EGF-like domains | |
| 3 | View Details | [526..665] | 10.93 | Hybrid domain of integrin beta; Integrin beta A domain; Integrin beta tail domain; Integrin beta EGF-like domains | |
| 4 | View Details | [666..728] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [689..728] | 1.002996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)