













| General Information: |
|
| Name(s) found: |
MED14_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1082 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTTIGSPQM LANEERLSNE MHALKNRSEQ NGQEQQGPVK NTQLHGPSAT DPETTATQKE 60
61 SLEMVPKDTS AATMTSAPPP ALPHVEINQV SLALVIRNLT VFTMKELAQY MKTNVHTQAN 120
121 EPNSAKKIRF LQLIIFLRTQ FLKLYVLVKW TRTIKQNNFH VLIDLLNWFR TTNMNVNNCI 180
181 WALKSSLNSM TNAKLPNVDL VTALEVLSLG RPNLPTHNFK LSGVSNSMDM VDGMAKVPIG 240
241 LILQRLKDLN LTVSIKIALM NIPKPLNSYH IKNGRIYFTV PNEFEIQLST VNRQSPLFFV 300
301 DLKLLFNTEA EQTVSAVTEA TSTNGDSENN EENSSSNGNN LPLNKPRLEK LINEILLKSN 360
361 DPLLSLYNFL HKYVLTLQLY MVHREFLKLA NGGKFSKSNL IHNYDSKKST ITVRYWLNGK 420
421 MDSKGKITIG IQRTTESLIL KWDNQSASRA KNMPVIYNNI VSNIEGILDE IMFNHARIIR 480
481 SELLARDIFQ EDEENSDVLL FQLPTTCVSM APIQLKIDLL SGQFYFRNPT PLLSNYASKI 540
541 NRAEGPEELA RILQQLKLDK IIHVLTTMFE NTGWSCSRII KIDKPIRTQV NTGGESVVKK 600
601 EDNKYAIAGN STTNSDVSLL LQRDLFIRLP HWPLNWYLIL SIISSKTSCV VEKRIGKIVS 660
661 QRGKWNLKYL DNSNVMTVKL ESITYQKIMI LQRTILNRII NHMLIDSLNQ LEIRNKICSS 720
721 EMINEQKLPQ YIIQGSNTND NISIITLELE SFLEGSKALN SILESSMFLR IDYSNSQIRL 780
781 YAKFKRNTMM IQCQIDKLYI HFVQEEPLAF YLEESFTNLG IIVQYLTKFR QKLMQLVVLT 840
841 DVVERLHKNF ESENFKIIAL QPNEISFKYL SNNDEDDKDC TIKISTNDDS IKNLTVQLSP 900
901 SNPQHIIQPF LDNSKMDYHF IFSYLQFTSS LFKALKVILN ERGGKFHESG SQYSTMVNIG 960
961 LHNLNEYQIV YYNPQAGTKI TICIELKTVL HNGRDKIQFH IHFADVAHIT TKSPAYPMMH1020
1021 QVRNQVFMLD TKRLGTPESV KPANASHAIR LGNGVACDPS EIEPILMEIH NILKVDSNSS1080
1081 SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
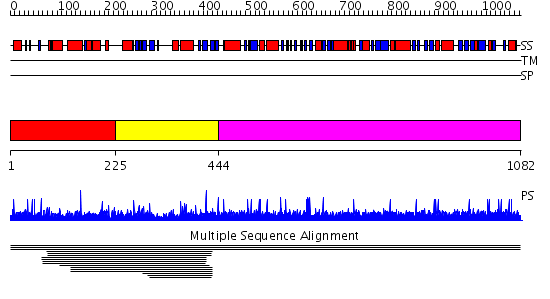
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..224] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [225..443] | 2.008975 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [444..1082] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)