













| General Information: |
|
| Name(s) found: |
MCM10_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDPREILAV DPYNNITSDE EDEQAIAREL EFMERKRQAL VERLKRKQEF KKPQDPNFEA 60
61 IEVPQSPTKN RVKVGSHNAT QQGTKFEGSN INEVRLSQLQ QQPKPPASTT TYFMEKFQNA 120
121 KKNEDKQIAK FESMMNARVH TFSTDEKKYV PIITNELESF SNLWVKKRYI PEDDLKRALH 180
181 EIKILRLGKL FAKIRPPKFQ EPEYANWATV GLISHKSDIK FTSSEKPVKF FMFTITDFQH 240
241 TLDVYIFGKK GVERYYNLRL GDVIAILNPE VLPWRPSGRG NFIKSFNLRI SHDFKCILEI 300
301 GSSRDLGWCP IVNKKTHKKC GSPINISLHK CCDYHREVQF RGTSAKRIEL NGGYALGAPT 360
361 KVDSQPSLYK AKGENGFNII KGTRKRLSEE EERLKKSSHN FTNSNSAKAF FDEKFQNPDM 420
421 LANLDNKRRK IIETKKSTAL SRELGKIMRR RESSGLEDKS VGERQKMKRT TESALQTGLI 480
481 QRLGFDPTHG KISQVLKSSV SGSEPKNNLL GKKKTVINDL LHYKKEKVIL APSKNEWFKK 540
541 RSHREEVWQK HFGSKETKET SDGSASDLEI I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
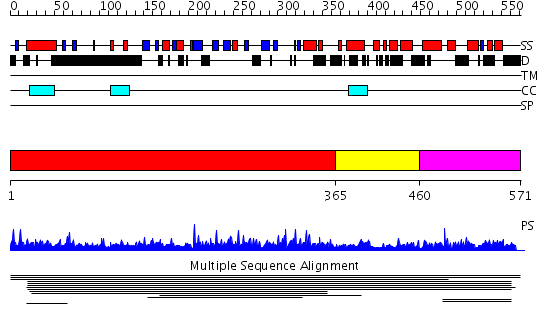
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..364] | 2.008917 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [365..459] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [460..571] | 1.005982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [487..571] | 2.016996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)