













| General Information: |
|
| Name(s) found: |
LCB4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVQKKLRAI LTDEGVLIKS QSHHMFNKHG QLRSGDSLSL LSCLSCLDDG TLSSDGGSFD 60
61 EDDSLELLPL NTTIPFNRIL NAKYVNVGQK GFNNGKISSN PFQTENLSSS SENDDVENHS 120
121 LSNDKAPVSE SQSFPKKDKW DTKTNTVKVS PDDSQDNSPS LGIKDNQQLI ELTFAVPKGH 180
181 DVIPQKLTLL IDHVSRKSRA NTGEENISSG TVEEILEKSY ENSKRNRSIL VIINPHGGKG 240
241 TAKNLFLTKA RPILVESGCK IEIAYTKYAR HAIDIAKDLD ISKYDTIACA SGDGIPYEVI 300
301 NGLYRRPDRV DAFNKLAVTQ LPCGSGNAMS ISCHWTNNPS YAALCLVKSI ETRIDLMCCS 360
361 QPSYMNEWPR LSFLSQTYGV IAESDINTEF IRWMGPVRFN LGVAFNIIQG KKYPCEVFVK 420
421 YAAKSKKELK VHFLENKDKN KGCLTFEPNP SPNSSPDLLS KNNINNSTKD ELSPNFLNED 480
481 NFKLKYPMTE PVPRDWEKMD SELTDNLTIF YTGKMPYIAK DTKFFPAALP ADGTIDLVIT 540
541 DARIPVTRMT PILLSLDKGS HVLEPEVIHS KILAYKIIPK VESGLFSVDG EKFPLEPLQV 600
601 EIMPMLCKTL LRNGRYIDTE FESM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
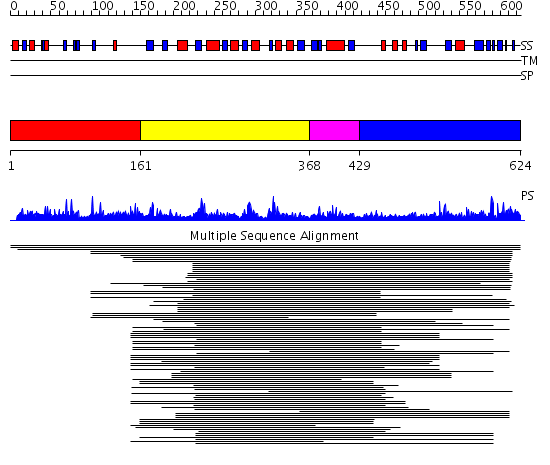
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [161..367] | 7.4 | Glycerol dehydrogenase | |
| 3 | View Details | [368..428] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [429..624] | 3.106994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)